Phosphatidylinositol 3,4,5-trisphosphate activity probes for the labeling and proteomic characterization of protein binding partners
- PMID: 22074223
- PMCID: PMC3649876
- DOI: 10.1021/bi201636s
Phosphatidylinositol 3,4,5-trisphosphate activity probes for the labeling and proteomic characterization of protein binding partners
Abstract
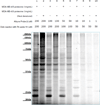
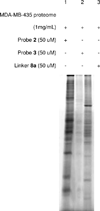
Phosphatidylinositol polyphosphate lipids, such as phosphatidylinositol 3,4,5-trisphosphate [PI(3,4,5)P₃], regulate critical biological processes, many of which are aberrant in disease. These lipids often act as site-specific ligands in interactions that enforce membrane association of protein binding partners. Herein, we describe the development of bifunctional activity probes corresponding to the headgroup of PI(3,4,5)P₃ that are effective for identifying and characterizing protein binding partners from complex samples, namely cancer cell extracts. These probes contain both a photoaffinity tag for covalent labeling of target proteins and a secondary handle for subsequent detection or manipulation of labeled proteins. Probes bearing different secondary tags were exploited, either by direct attachment of a fluorescent dye for optical detection or by using an alkyne that can be derivatized after protein labeling via click chemistry. First, we describe the design and modular synthetic strategy used to generate multiple probes with different reporter tags of use for characterizing probe-labeled proteins. Next, we report initial labeling studies using purified protein, the PH domain of Akt, in which probes were found to label this target, as judged by in-gel detection. Furthermore, protein labeling was abrogated by controls including competition with an unlabeled PI(3,4,5)P₃ headgroup analogue as well as through protein denaturation, indicating specific labeling. In addition, probes featuring linkers of different lengths between the PI(3,4,5)P₃ headgroup and photoaffinity tag led to variations in protein labeling, indicating that a shorter linker was more effective in this case. Finally, proteomic labeling studies were performed using cell extracts; labeled proteins were observed by in-gel detection and characterized using postlabeling with biotin, affinity chromatography, and identification via tandem mass spectrometry. These studies yielded a total of 265 proteins, including both known and novel candidate PI(3,4,5)P₃-binding proteins.
Figures
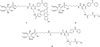





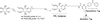

Similar articles
-
Development of clickable active site-directed photoaffinity probes for γ-secretase.Bioorg Med Chem Lett. 2012 Apr 15;22(8):2997-3000. doi: 10.1016/j.bmcl.2012.02.027. Epub 2012 Feb 20. Bioorg Med Chem Lett. 2012. PMID: 22418280 Free PMC article.
-
Recent developments and applications of clickable photoprobes in medicinal chemistry and chemical biology.Future Med Chem. 2015;7(16):2143-71. doi: 10.4155/fmc.15.136. Epub 2015 Oct 29. Future Med Chem. 2015. PMID: 26511756 Review.
-
Modular synthesis of biologically active phosphatidic acid probes using click chemistry.Mol Biosyst. 2009 Sep;5(9):962-72. doi: 10.1039/b901420a. Epub 2009 May 7. Mol Biosyst. 2009. PMID: 19668861 Free PMC article.
-
Exploiting bioorthogonal chemistry to elucidate protein-lipid binding interactions and other biological roles of phospholipids.Acc Chem Res. 2011 Sep 20;44(9):686-98. doi: 10.1021/ar200060y. Epub 2011 May 6. Acc Chem Res. 2011. PMID: 21548554
-
Photoaffinity labeling in drug discovery and developments: chemical gateway for entering proteomic frontier.Curr Top Med Chem. 2002 Mar;2(3):271-88. doi: 10.2174/1568026023394182. Curr Top Med Chem. 2002. PMID: 11944820 Review.
Cited by
-
When PIP2 Meets p53: Nuclear Phosphoinositide Signaling in the DNA Damage Response.Front Cell Dev Biol. 2022 May 13;10:903994. doi: 10.3389/fcell.2022.903994. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35646908 Free PMC article. Review.
-
Cellular and molecular interactions of phosphoinositides and peripheral proteins.Chem Phys Lipids. 2014 Sep;182:3-18. doi: 10.1016/j.chemphyslip.2014.02.002. Epub 2014 Feb 17. Chem Phys Lipids. 2014. PMID: 24556335 Free PMC article. Review.
-
Photoaffinity labelling strategies for mapping the small molecule-protein interactome.Org Biomol Chem. 2021 Sep 22;19(36):7792-7809. doi: 10.1039/d1ob01353j. Org Biomol Chem. 2021. PMID: 34549230 Free PMC article. Review.
-
Synthetic Lipid Biology.Chem Rev. 2025 Feb 26;125(4):2502-2560. doi: 10.1021/acs.chemrev.4c00761. Epub 2025 Jan 13. Chem Rev. 2025. PMID: 39805091 Review.
-
Modular Approaches to Synthesize Activity- and Affinity-Based Chemical Probes.Front Chem. 2021 Apr 15;9:644811. doi: 10.3389/fchem.2021.644811. eCollection 2021. Front Chem. 2021. PMID: 33937194 Free PMC article. Review.
References
-
- Best MD, Zhang HL, Prestwich GD. Inositol polyphosphates, diphosphoinositol polyphosphates and phosphatidylinositol polyphosphate lipids: Structure, synthesis, and development of probes for studying biological activity. Nat. Prod. Rep. 2010;27:1403–1430. - PubMed
-
- Conway SJ, Miller GJ. Biology-enabling inositol phosphates, phosphatidylinositol phosphates and derivatives. Nat. Prod. Rep. 2007;24:687–707. - PubMed
-
- Lemmon MA. Membrane recognition by phospholipid-binding domains. Nat. Rev. Mol. Cell Biol. 2008;9:99–111. - PubMed
-
- Cho W, Stahelin RV. Membrane-protein interactions in cell signaling and membrane trafficking. Annu. Rev. Biophys. Biomol. Struct. 2005;34:119–151. - PubMed
-
- Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

