Research resource: interactome of human embryo implantation: identification of gene expression pathways, regulation, and integrated regulatory networks
- PMID: 22074951
- PMCID: PMC5417164
- DOI: 10.1210/me.2011-1196
Research resource: interactome of human embryo implantation: identification of gene expression pathways, regulation, and integrated regulatory networks
Abstract
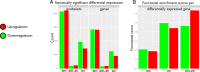
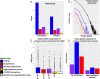
A prerequisite for successful embryo implantation is adequate preparation of receptive endometrium and the establishment and maintenance of a viable embryo. The success of implantation further relies upon a two-way dialogue between the embryo and uterus. However, molecular bases of these preimplantation and implantation processes in humans are not well known. We performed genome expression analyses of human embryos (n = 128) and human endometria (n = 8). We integrated these data with protein-protein interactions in order to identify molecular networks within the endometrium and the embryo, and potential embryo-endometrium interactions at the time of implantation. For that, we applied a novel network profiling algorithm HyperModules, which combines topological module identification and functional enrichment analysis. We found a major wave of transcriptional down-regulation in preimplantation embryos. In receptive-stage endometrium, several genes and signaling pathways were identified, including JAK-STAT signaling and inflammatory pathways. The main curated embryo-endometrium interaction network highlighted the importance of cell adhesion molecules in the implantation process. We also identified cytokine-cytokine receptor interactions involved in implantation, where osteopontin (SPP1), leukemia inhibitory factor (LIF) and leptin (LEP) pathways were intertwining. Further, we identified a number of novel players in human embryo-endometrium interactions, such as apolipoprotein D (APOD), endothelin 1 (END1), fibroblast growth factor 7 (FGF7), gastrin (GAST), kringle containing trnasmembrane protein 1 (KREMEN1), neuropilin 1 (NRP1), serpin peptidase inhibitor clade A member 3 (SERPINA3), versican (VCAN), and others. Our findings provide a fundamental resource for better understanding of the genetic network that leads to successful embryo implantation. We demonstrate the first systems biology approach into the complex molecular network of the implantation process in humans.
Figures



Comment in
-
Editorial: molecular endocrinology articles in the spotlight for January 2012.Mol Endocrinol. 2012 Jan;26(1):1. doi: 10.1210/me.2011-1345. Mol Endocrinol. 2012. PMID: 22210759 Free PMC article. No abstract available.
References
-
- Giudice LC. 1999. Genes associated with embryonic attachment and implantation and the role of progesterone. J Reprod Med 44:165–171 - PubMed
-
- Harper MJ. 1992. The implantation window. Baillieres Clin Obstet Gynaecol 6:351–371 - PubMed
-
- Wilcox AJ , Baird DD , Weinberg CR. 1999. Time of implantation of the conceptus and loss of pregnancy. N Engl J Med 340:1796–1799 - PubMed
-
- Carson DD , Bagchi I , Dey SK , Enders AC , Fazleabas AT , Lessey BA , Yoshinaga K. 2000. Embryo implantation. Dev Biol 223:217–237 - PubMed
-
- Giudice LC. 1999. Potential biochemical markers of uterine receptivity. Hum Reprod 14(Suppl 2):3–16 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

