Probing the oligomeric state and interaction surfaces of Fukutin-I in dilauroylphosphatidylcholine bilayers
- PMID: 22075563
- PMCID: PMC3269570
- DOI: 10.1007/s00249-011-0773-5
Probing the oligomeric state and interaction surfaces of Fukutin-I in dilauroylphosphatidylcholine bilayers
Abstract
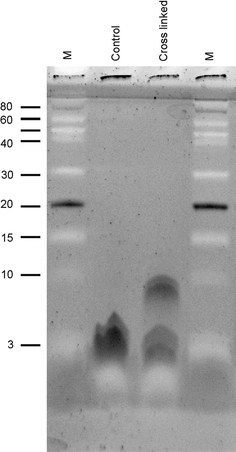
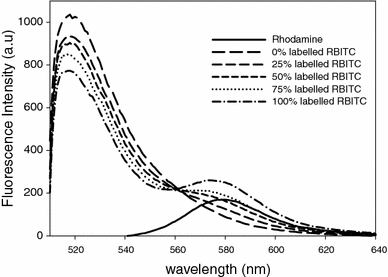
Fukutin-I is localised to the endoplasmic reticulum or Golgi apparatus within the cell, where it is believed to function as a glycosyltransferase. Its localisation within the cell is thought to to be mediated by the interaction of its N-terminal transmembrane domain with the lipid bilayers surrounding these compartments, each of which possesses a distinctive lipid composition. However, it remains unclear at the molecular level how the interaction between the transmembrane domains of this protein and the surrounding lipid bilayer drives its retention within these compartments. In this work, we employed chemical cross-linking and fluorescence resonance energy transfer measurements in conjunction with multiscale molecular dynamics simulations to determine the oligomeric state of the protein within dilauroylphosphatidylcholine bilayers to identify interactions between the transmembrane domains and to ascertain any role these interactions may play in protein localisation. Our studies reveal that the N-terminal transmembrane domain of Fukutin-I exists as dimer within dilauroylphosphatidylcholine bilayers and that this interaction is driven by interactions between a characteristic TXXSS motif. Furthermore residues close to the N-terminus that have previously been shown to play a key role in the clustering of lipids are shown to also play a major role in anchoring the protein in the membrane.
Figures



Similar articles
-
Stability and membrane orientation of the fukutin transmembrane domain: a combined multiscale molecular dynamics and circular dichroism study.Biochemistry. 2010 Dec 28;49(51):10796-802. doi: 10.1021/bi101743w. Epub 2010 Dec 6. Biochemistry. 2010. PMID: 21105749 Free PMC article.
-
Potential of mean force analysis of the self-association of leucine-rich transmembrane α-helices: difference between atomistic and coarse-grained simulations.J Chem Phys. 2014 Aug 21;141(7):075101. doi: 10.1063/1.4891932. J Chem Phys. 2014. PMID: 25149815
-
The transmembrane domains of the bacterial cell division proteins FtsB and FtsL form a stable high-order oligomer.Biochemistry. 2013 Oct 29;52(43):7542-50. doi: 10.1021/bi4009837. Epub 2013 Oct 18. Biochemistry. 2013. PMID: 24083359 Free PMC article.
-
Lipid-protein interplay and lateral organization in biomembranes.Chem Phys Lipids. 2015 Jul;189:48-55. doi: 10.1016/j.chemphyslip.2015.05.008. Epub 2015 May 30. Chem Phys Lipids. 2015. PMID: 26036778 Review.
-
The influence of cholesterol on membrane protein structure, function, and dynamics studied by molecular dynamics simulations.Biochim Biophys Acta. 2015 Sep;1848(9):1783-95. doi: 10.1016/j.bbamem.2015.03.029. Epub 2015 Apr 1. Biochim Biophys Acta. 2015. PMID: 25839353 Review.
Cited by
-
CHARMM-GUI PACE CG Builder for solution, micelle, and bilayer coarse-grained simulations.J Chem Inf Model. 2014 Mar 24;54(3):1003-9. doi: 10.1021/ci500007n. Epub 2014 Mar 13. J Chem Inf Model. 2014. PMID: 24624945 Free PMC article.
-
In pursuit of an accurate spatial and temporal model of biomolecules at the atomistic level: a perspective on computer simulation.Acta Crystallogr D Biol Crystallogr. 2015 Jan 1;71(Pt 1):162-72. doi: 10.1107/S1399004714026777. Epub 2015 Jan 1. Acta Crystallogr D Biol Crystallogr. 2015. PMID: 25615870 Free PMC article.
-
Helical membrane protein conformations and their environment.Eur Biophys J. 2013 Oct;42(10):731-55. doi: 10.1007/s00249-013-0925-x. Epub 2013 Sep 1. Eur Biophys J. 2013. PMID: 23996195 Free PMC article. Review.
-
High degree of conservation of the enzymes synthesizing the laminin-binding glycoepitope of α-dystroglycan.Open Biol. 2021 Sep;11(9):210104. doi: 10.1098/rsob.210104. Epub 2021 Sep 29. Open Biol. 2021. PMID: 34582712 Free PMC article.
-
Interaction between the NS4B amphipathic helix, AH2, and charged lipid headgroups alters membrane morphology and AH2 oligomeric state--Implications for the Hepatitis C virus life cycle.Biochim Biophys Acta. 2015 Aug;1848(8):1671-7. doi: 10.1016/j.bbamem.2015.04.015. Epub 2015 May 2. Biochim Biophys Acta. 2015. PMID: 25944559 Free PMC article.
References
-
- Berendsen HJC, Postma JPM, van Gunsteren WF, Hermans J. In intermolecular forces. Dordrecht: Reidel; 1981.
-
- Berendsen H, van der Spoel D, van Drunen R (1995) GROMACS—a message passing parallel molecular-dynamics implementation. Comput Phys Comm 91:43–56
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

