Interconversion between intestinal stem cell populations in distinct niches
- PMID: 22075725
- PMCID: PMC3705713
- DOI: 10.1126/science.1213214
Interconversion between intestinal stem cell populations in distinct niches
Abstract
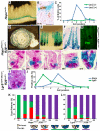
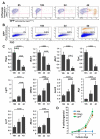
Intestinal epithelial stem cell identity and location have been the subject of substantial research. Cells in the +4 niche are slow-cycling and label-retaining, whereas a different stem cell niche located at the crypt base is occupied by crypt base columnar (CBC) cells. CBCs are distinct from +4 cells, and the relationship between them is unknown, though both give rise to all intestinal epithelial lineages. We demonstrate that Hopx, an atypical homeobox protein, is a specific marker of +4 cells. Hopx-expressing cells give rise to CBCs and all mature intestinal epithelial lineages. Conversely, CBCs can give rise to +4 Hopx-positive cells. These findings demonstrate a bidirectional lineage relationship between active and quiescent stem cells in their niches.
Figures


Comment in
-
Tales from the crypt: the expanding role of slow cycling intestinal stem cells.Cell Stem Cell. 2012 Jan 6;10(1):2-4. doi: 10.1016/j.stem.2011.12.012. Cell Stem Cell. 2012. PMID: 22226346 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

