An in vivo polymicrobial biofilm wound infection model to study interspecies interactions
- PMID: 22076151
- PMCID: PMC3208625
- DOI: 10.1371/journal.pone.0027317
An in vivo polymicrobial biofilm wound infection model to study interspecies interactions
Abstract
Chronic wound infections are typically polymicrobial; however, most in vivo studies have focused on monospecies infections. This project was designed to develop an in vivo, polymicrobial, biofilm-related, infected wound model in order to study multispecies biofilm dynamics and in relation to wound chronicity. Multispecies biofilms consisting of both Gram negative and Gram positive strains, as well as aerobes and anaerobes, were grown in vitro and then transplanted onto the wounds of mice. These in vitro-to-in vivo multi-species biofilm transplants generated polymicrobial wound infections, which remained heterogeneous with four bacterial species throughout the experiment. We observed that wounded mice given multispecies biofilm infections displayed a wound healing impairment over mice infected with a single-species of bacteria. In addition, the bacteria in the polymicrobial wound infections displayed increased antimicrobial tolerance in comparison to those in single species infections. These data suggest that synergistic interactions between different bacterial species in wounds may contribute to healing delays and/or antibiotic tolerance.
Conflict of interest statement
Figures






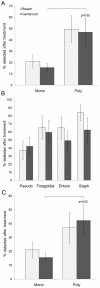
References
-
- American Burn Association website. Burn Incidence and Treatment in the US: 2007 Fact Sheet. Available: http://www.ameriburn.org/resources_factsheet.php. Accessed 2011 Oct 17.
-
- Center for Disease Control. . Department of Health and Human Services, Centers for Disease Control and Prevention (Eds.) Atlanta, GA: 2002. National estimates and general information on diabetes in the U.S. Revised edition in:Center U.S.
-
- National Institute of General Medical Sciences website. Fact Sheet:Trauma, Shock, Burn and Injury: Fact and Figures. 2007;17 http://www.nigms.nih.gov/Education/Factsheet_Trauma.htm. Accessed 2011 Oct.
-
- James GA, Swogger E, Wolcott R, Pulcini E, Secor P, et al. Biofilms in chronic wounds. Wound Repair Regen. 2008;16:37–44. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

