Multifunctional proteins revealed by overlapping clustering in protein interaction network
- PMID: 22080466
- PMCID: PMC3244771
- DOI: 10.1093/bioinformatics/btr621
Multifunctional proteins revealed by overlapping clustering in protein interaction network
Abstract
Motivation: Multifunctional proteins perform several functions. They are expected to interact specifically with distinct sets of partners, simultaneously or not, depending on the function performed. Current graph clustering methods usually allow a protein to belong to only one cluster, therefore impeding a realistic assignment of multifunctional proteins to clusters.
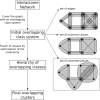
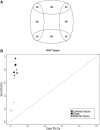
Results: Here, we present Overlapping Cluster Generator (OCG), a novel clustering method which decomposes a network into overlapping clusters and which is, therefore, capable of correct assignment of multifunctional proteins. The principle of OCG is to cover the graph with initial overlapping classes that are iteratively fused into a hierarchy according to an extension of Newman's modularity function. By applying OCG to a human protein-protein interaction network, we show that multifunctional proteins are revealed at the intersection of clusters and demonstrate that the method outperforms other existing methods on simulated graphs and PPI networks.
Availability: This software can be downloaded from http://tagc.univ-mrs.fr/welcome/spip.php?rubrique197
Contact: brun@tagc.univ-mrs.fr
Supplementary information: Supplementary data are available at Bioinformatics online.
Figures


References
-
- Adamcsek B., et al. CFinder: locating cliques and overlapping modules in biological networks. Bioinformatics. 2006;22:1021–1023. - PubMed
-
- Ahn Y.-Y., et al. Link communities reveal multiscale complexity in networks. Nature. 2010;466:761–764. - PubMed
-
- Aittokallio T., Schwikowski B. Graph-based methods for analysing networks in cell biology. Brief. Bioinform. 2006;7:243–255. - PubMed
-
- Angelelli J.B., Reboul L. Proceedings of JOBIM 2008. Lille: University Press; 2008. Network modularity optimization by a fusion-fission process and application to protein-protein interactions networks; pp. 105–110.
-
- Ahn J., et al. Integrative gene network construction for predicting a set of complementary prostate cancer genes. Bioinformatics. 2011;27:1846–1853. - PubMed

