HotRegion: a database of predicted hot spot clusters
- PMID: 22080558
- PMCID: PMC3245113
- DOI: 10.1093/nar/gkr929
HotRegion: a database of predicted hot spot clusters
Abstract
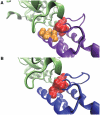
Hot spots are energetically important residues at protein interfaces and they are not randomly distributed across the interface but rather clustered. These clustered hot spots form hot regions. Hot regions are important for the stability of protein complexes, as well as providing specificity to binding sites. We propose a database called HotRegion, which provides the hot region information of the interfaces by using predicted hot spot residues, and structural properties of these interface residues such as pair potentials of interface residues, accessible surface area (ASA) and relative ASA values of interface residues of both monomer and complex forms of proteins. Also, the 3D visualization of the interface and interactions among hot spot residues are provided. HotRegion is accessible at http://prism.ccbb.ku.edu.tr/hotregion.
Figures
Similar articles
-
HotPoint: hot spot prediction server for protein interfaces.Nucleic Acids Res. 2010 Jul;38(Web Server issue):W402-6. doi: 10.1093/nar/gkq323. Epub 2010 May 5. Nucleic Acids Res. 2010. PMID: 20444871 Free PMC article.
-
HotSprint: database of computational hot spots in protein interfaces.Nucleic Acids Res. 2008 Jan;36(Database issue):D662-6. doi: 10.1093/nar/gkm813. Epub 2007 Oct 24. Nucleic Acids Res. 2008. PMID: 17959648 Free PMC article.
-
Identification of computational hot spots in protein interfaces: combining solvent accessibility and inter-residue potentials improves the accuracy.Bioinformatics. 2009 Jun 15;25(12):1513-20. doi: 10.1093/bioinformatics/btp240. Epub 2009 Apr 8. Bioinformatics. 2009. PMID: 19357097
-
Anatomy of hot spots in protein interfaces.J Mol Biol. 1998 Jul 3;280(1):1-9. doi: 10.1006/jmbi.1998.1843. J Mol Biol. 1998. PMID: 9653027 Review.
-
Machine Learning Approaches for Protein⁻Protein Interaction Hot Spot Prediction: Progress and Comparative Assessment.Molecules. 2018 Oct 4;23(10):2535. doi: 10.3390/molecules23102535. Molecules. 2018. PMID: 30287797 Free PMC article. Review.
Cited by
-
Computational Design of Hypothetical New Peptides Based on a Cyclotide Scaffold as HIV gp120 Inhibitor.PLoS One. 2015 Oct 30;10(11):e0139562. doi: 10.1371/journal.pone.0139562. eCollection 2015. PLoS One. 2015. PMID: 26517259 Free PMC article.
-
Enriching Traditional Protein-protein Interaction Networks with Alternative Conformations of Proteins.Sci Rep. 2017 Aug 3;7(1):7180. doi: 10.1038/s41598-017-07351-0. Sci Rep. 2017. PMID: 28775330 Free PMC article.
-
Design of novel peptide inhibitors against the conserved bacterial transcription terminator, Rho.J Biol Chem. 2021 Jan-Jun;296:100653. doi: 10.1016/j.jbc.2021.100653. Epub 2021 May 15. J Biol Chem. 2021. PMID: 33845047 Free PMC article.
-
Interolog interfaces in protein-protein docking.Proteins. 2015 Nov;83(11):1940-6. doi: 10.1002/prot.24788. Epub 2015 Sep 29. Proteins. 2015. PMID: 25740680 Free PMC article.
-
PPInS: a repository of protein-protein interaction sitesbase.Sci Rep. 2018 Aug 20;8(1):12453. doi: 10.1038/s41598-018-30999-1. Sci Rep. 2018. PMID: 30127348 Free PMC article.
References
-
- Illingworth CJ, Scott PD, Parkes KE, Snell CR, Campbell MP, Reynolds CA. Connectivity and binding-site recognition: applications relevant to drug design. J. Comput. Chem. 2010;31:2677–2688. - PubMed
-
- Kim PM, Lu LJ, Xia Y, Gerstein MB. Relating three-dimensional structures to protein networks provides evolutionary insights. Science. 2006;314:1938–1941. - PubMed
-
- Keskin O, Nussinov R. Similar binding sites and different partners: implications to shared proteins in cellular pathways. Structure. 2007;15:341–354. - PubMed