Discovery of active enhancers through bidirectional expression of short transcripts
- PMID: 22082242
- PMCID: PMC3334599
- DOI: 10.1186/gb-2011-12-11-r113
Discovery of active enhancers through bidirectional expression of short transcripts
Abstract
Background: Long-range regulatory elements, such as enhancers, exert substantial control over tissue-specific gene expression patterns. Genome-wide discovery of functional enhancers in different cell types is important for our understanding of genome function as well as human disease etiology.
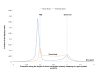
Results: In this study, we developed an in silico approach to model the previously reported phenomenon of transcriptional pausing, accompanied by divergent transcription, at active promoters. We then used this model for large-scale prediction of non-promoter-associated bidirectional expression of short transcripts. Our predictions were significantly enriched for DNase hypersensitive sites, histone H3 lysine 27 acetylation (H3K27ac), and other chromatin marks associated with active rather than poised or repressed enhancers. We also detected modest bidirectional expression at binding sites of the CCCTC-factor (CTCF) genome-wide, particularly those that overlap H3K27ac.
Conclusions: Our findings indicate that the signature of bidirectional expression of short transcripts, learned from promoter-proximal transcriptional pausing, can be used to predict active long-range regulatory elements genome-wide, likely due in part to specific association of RNA polymerase with enhancer regions.
Figures



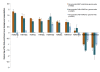

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

