Resveratrol-mediated autophagy requires WIPI-1-regulated LC3 lipidation in the absence of induced phagophore formation
- PMID: 22082875
- PMCID: PMC3288019
- DOI: 10.4161/auto.7.12.17802
Resveratrol-mediated autophagy requires WIPI-1-regulated LC3 lipidation in the absence of induced phagophore formation
Abstract
Canonical autophagy is positively regulated by the Beclin 1/phosphatidylinositol 3-kinase class III (PtdIns3KC3) complex that generates an essential phospholipid, phosphatidylinositol 3-phosphate (PtdIns(3)P), for the formation of autophagosomes. Previously, we identified the human WIPI protein family and found that WIPI-1 specifically binds PtdIns(3)P, accumulates at the phagophore and becomes a membrane protein of generated autophagosomes. Combining siRNA-mediated protein downregulation with automated high through-put analysis of PtdIns(3)P-dependent autophagosomal membrane localization of WIPI-1, we found that WIPI-1 functions upstream of both Atg7 and Atg5, and stimulates an increase of LC3-II upon nutrient starvation. Resveratrol-mediated autophagy was shown to enter autophagic degradation in a noncanonical manner, independent of Beclin 1 but dependent on Atg7 and Atg5. By using electron microscopy, LC3 lipidation and GFP-LC3 puncta-formation assays we confirmed these results and found that this effect is partially wortmannin-insensitive. In line with this, resveratrol did not promote phagophore localization of WIPI-1, WIPI-2 or the Atg16L complex above basal level. In fact, the presence of resveratrol in nutrient-free conditions inhibited phagophore localization of WIPI-1. Nevertheless, we found that resveratrol-mediated autophagy functionally depends on canonical-driven LC3-II production, as shown by siRNA-mediated downregulation of WIPI-1 or WIPI-2. From this it is tempting to speculate that resveratrol promotes noncanonical autophagic degradation downstream of the PtdIns(3)P-WIPI-Atg7-Atg5 pathway, by engaging a distinct subset of LC3-II that might be generated at membrane origins apart from canonical phagophore structures.
Figures



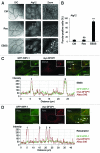
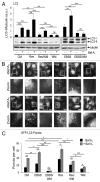
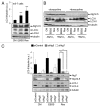

Similar articles
-
Ca2+/calmodulin-dependent kinase (CaMK) signaling via CaMKI and AMP-activated protein kinase contributes to the regulation of WIPI-1 at the onset of autophagy.Mol Pharmacol. 2011 Dec;80(6):1066-75. doi: 10.1124/mol.111.071761. Epub 2011 Sep 6. Mol Pharmacol. 2011. PMID: 21896713
-
Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation.Autophagy. 2010 May;6(4):506-22. doi: 10.4161/auto.6.4.11863. Epub 2010 May 16. Autophagy. 2010. PMID: 20505359
-
Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins.Autophagy. 2010 Aug;6(6):764-76. doi: 10.4161/auto.6.6.12709. Autophagy. 2010. PMID: 20639694 Free PMC article.
-
WIPI proteins: essential PtdIns3P effectors at the nascent autophagosome.J Cell Sci. 2015 Jan 15;128(2):207-17. doi: 10.1242/jcs.146258. J Cell Sci. 2015. PMID: 25568150 Review.
-
Molecular basis of canonical and bactericidal autophagy.Int Immunol. 2009 Nov;21(11):1199-204. doi: 10.1093/intimm/dxp088. Epub 2009 Sep 7. Int Immunol. 2009. PMID: 19737785 Review.
Cited by
-
WIPI-Mediated Autophagy and Longevity.Cells. 2015 May 22;4(2):202-17. doi: 10.3390/cells4020202. Cells. 2015. PMID: 26010754 Free PMC article. Review.
-
Analysis of the Compositional Features and Codon Usage Pattern of Genes Involved in Human Autophagy.Cells. 2022 Oct 12;11(20):3203. doi: 10.3390/cells11203203. Cells. 2022. PMID: 36291071 Free PMC article.
-
Estrogen receptor α regulates non-canonical autophagy that provides stress resistance to neuroblastoma and breast cancer cells and involves BAG3 function.Cell Death Dis. 2015 Jul 9;6(7):e1812. doi: 10.1038/cddis.2015.181. Cell Death Dis. 2015. PMID: 26158518 Free PMC article.
-
Akt/mTOR-Mediated Autophagy Confers Resistance To BET Inhibitor JQ1 In Ovarian Cancer.Onco Targets Ther. 2019 Oct 3;12:8063-8074. doi: 10.2147/OTT.S220267. eCollection 2019. Onco Targets Ther. 2019. PMID: 31632060 Free PMC article.
-
Differential regulatory functions of three classes of phosphatidylinositol and phosphoinositide 3-kinases in autophagy.Autophagy. 2015;11(10):1711-28. doi: 10.1080/15548627.2015.1043076. Autophagy. 2015. PMID: 26018563 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
