Predicting synthetic gene networks
- PMID: 22083736
- PMCID: PMC7120583
- DOI: 10.1007/978-1-61779-412-4_4
Predicting synthetic gene networks
Abstract
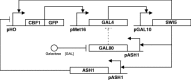
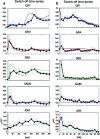
Synthetic biology aims at designing and building new biological functions in living organisms. The complexity of cellular regulation (regulatory, metabolic, and signaling interactions, and their coordinated action) can be tackled via the development of quantitative mathematical models. These models are useful to test biological hypotheses and observations, and to predict the possible behaviors of a synthetic network. Indeed, synthetic biology uses such models to design synthetic networks, prior to their construction in the cell, to perform specific tasks, or to change a biological process in a desired way. The synthetic network is built by assembling biological "parts" taken from different systems; therefore it is fundamental to identify, isolate, and test regulatory motifs which occur frequently in biological pathways. In this chapter, we describe how to model and predict the behavior of synthetic networks in two difference cases: (1) a synthetic network composed of five genes regulating each other through a variety of regulatory interactions in the yeast Saccharomyces cerevisiae (2) a synthetic transcriptional positive feedback loop stably integrated in Human Embryonic Kidney 293 cells (HEK293).
Figures




References
-
- Cuccato G. Systems and Synthetic biology: tackling genetic networks and complex diseases. Heredity. 2009;102:527–532. - PubMed
-
- Cantone I., Marucci L., Iorio F., Ricci M. A., Belcastro V., Bansal M., Santini S., Bernardo M., Bernardo D., Cosma M. P. A yeast synthetic network for in vivo assessment of reverse-engineering and modeling approaches. Cell. 2009;137:172–181. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

