An entirely specific type I A-kinase anchoring protein that can sequester two molecules of protein kinase A at mitochondria
- PMID: 22084075
- PMCID: PMC3228425
- DOI: 10.1073/pnas.1107182108
An entirely specific type I A-kinase anchoring protein that can sequester two molecules of protein kinase A at mitochondria
Abstract
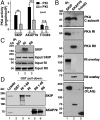
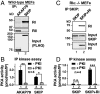
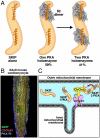
A-kinase anchoring proteins (AKAPs) tether the cAMP-dependent protein kinase (PKA) to intracellular sites where they preferentially phosphorylate target substrates. Most AKAPs exhibit nanomolar affinity for the regulatory (RII) subunit of the type II PKA holoenzyme, whereas dual-specificity anchoring proteins also bind the type I (RI) regulatory subunit of PKA with 10-100-fold lower affinity. A range of cellular, biochemical, biophysical, and genetic approaches comprehensively establish that sphingosine kinase interacting protein (SKIP) is a truly type I-specific AKAP. Mapping studies located anchoring sites between residues 925-949 and 1,140-1,175 of SKIP that bind RI with dissociation constants of 73 and 774 nM, respectively. Molecular modeling and site-directed mutagenesis approaches identify Phe 929 and Tyr 1,151 as RI-selective binding determinants in each anchoring site. SKIP complexes exist in different states of RI-occupancy as single-molecule pull-down photobleaching experiments show that 41 ± 10% of SKIP sequesters two YFP-RI dimers, whereas 59 ± 10% of the anchoring protein binds a single YFP-RI dimer. Imaging, proteomic analysis, and subcellular fractionation experiments reveal that SKIP is enriched at the inner mitochondrial membrane where it associates with a prominent PKA substrate, the coiled-coil helix protein ChChd3.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Keren H, Lev-Maor G, Ast G. Alternative splicing and evolution: Diversification, exon definition and function. Nat Rev Genet. 2010;11:345–355. - PubMed
-
- Hunter T. Protein kinases and phosphatases: The yin and yang of protein phosphorylation and signaling. Cell. 1995;80:225–236. - PubMed
-
- Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. - PubMed
-
- Mitchison T, Kirschner M. Dynamic instability of microtubule growth. Nature. 1984;312:237–242. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

