Temporally structured metapopulation dynamics and persistence of influenza A H3N2 virus in humans
- PMID: 22084096
- PMCID: PMC3228450
- DOI: 10.1073/pnas.1109314108
Temporally structured metapopulation dynamics and persistence of influenza A H3N2 virus in humans
Abstract
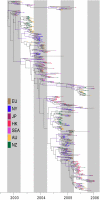
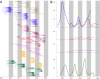
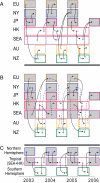
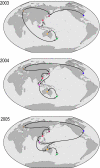
Populations of seasonal influenza virus experience strong annual bottlenecks that pose a considerable extinction risk. It has been suggested that an influenza source population located in tropical Southeast or East Asia seeds annual temperate epidemics. Here we investigate the seasonal dynamics and migration patterns of influenza A H3N2 virus by analysis of virus samples obtained from 2003 to 2006 from Australia, Europe, Japan, New York, New Zealand, Southeast Asia, and newly sequenced viruses from Hong Kong. In contrast to annual temperate epidemics, relatively low levels of relative genetic diversity and no seasonal fluctuations characterized virus populations in tropical Southeast Asia and Hong Kong. Bayesian phylogeographic analysis using discrete temporal and spatial characters reveal high rates of viral migration between urban centers tested. Although the virus population that migrated between Southeast Asia and Hong Kong persisted through time, this was dependent on virus input from temperate regions and these tropical regions did not maintain a source for annual H3N2 influenza epidemics. We further show that multiple lineages may seed annual influenza epidemics, and that each region may function as a potential source population. We therefore propose that the global persistence of H3N2 influenza A virus is the result of a migrating metapopulation in which multiple different localities may seed seasonal epidemics in temperate regions in a given year. Such complex global migration dynamics may confound control efforts and contribute to the emergence and spread of antigenic variants and drug-resistant viruses.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

