Evolution of networks and sequences in eukaryotic cell cycle control
- PMID: 22084380
- PMCID: PMC3203458
- DOI: 10.1098/rstb.2011.0078
Evolution of networks and sequences in eukaryotic cell cycle control
Abstract
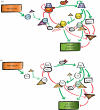
The molecular networks regulating the G1-S transition in budding yeast and mammals are strikingly similar in network structure. However, many of the individual proteins performing similar network roles appear to have unrelated amino acid sequences, suggesting either extremely rapid sequence evolution, or true polyphyly of proteins carrying out identical network roles. A yeast/mammal comparison suggests that network topology, and its associated dynamic properties, rather than regulatory proteins themselves may be the most important elements conserved through evolution. However, recent deep phylogenetic studies show that fungal and animal lineages are relatively closely related in the opisthokont branch of eukaryotes. The presence in plants of cell cycle regulators such as Rb, E2F and cyclins A and D, that appear lost in yeast, suggests cell cycle control in the last common ancestor of the eukaryotes was implemented with this set of regulatory proteins. Forward genetics in non-opisthokonts, such as plants or their green algal relatives, will provide direct information on cell cycle control in these organisms, and may elucidate the potentially more complex cell cycle control network of the last common eukaryotic ancestor.
Figures



References
-
- Lee M. G., Nurse P. 1987. Complementation used to clone a human homologue of the fission yeast cell cycle control gene cdc2. Nature 327, 31–35 10.1038/327031a0 (doi:10.1038/327031a0) - DOI - DOI - PubMed
-
- Forsburg S. L., Nurse P. 1991. Identification of a G1-type cyclin puc1+ in the fission yeast Schizosaccharomyces pombe. Nature 351, 245–248 10.1038/351245a0 (doi:10.1038/351245a0) - DOI - DOI - PubMed
-
- Koff A., Cross F., Fisher A., Schumacher J., Leguellec K., Philippe M., Roberts J. M. 1991. Human cyclin E, a new cyclin that interacts with two members of the CDC2 gene family. Cell 66, 1217–1228 10.1016/0092-8674(91)90044-Y (doi:10.1016/0092-8674(91)90044-Y) - DOI - DOI - PubMed
-
- Léopold P., O'Farrell P. H. 1991. An evolutionarily conserved cyclin homolog from Drosophila rescues yeast deficient in G1 cyclins. Cell 66, 1207–1216 10.1016/0092-8674(91)90043-X (doi:10.1016/0092-8674(91)90043-X) - DOI - DOI - PMC - PubMed
-
- Lew D. J., Dulić V., Reed S. I. 1991. Isolation of three novel human cyclins by rescue of G1 cyclin (Cln) function in yeast. Cell 66, 1197–1206 10.1016/0092-8674(91)90042-W (doi:10.1016/0092-8674(91)90042-W) - DOI - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

