Transcriptional regulation in Saccharomyces cerevisiae: transcription factor regulation and function, mechanisms of initiation, and roles of activators and coactivators
- PMID: 22084422
- PMCID: PMC3213380
- DOI: 10.1534/genetics.111.127019
Transcriptional regulation in Saccharomyces cerevisiae: transcription factor regulation and function, mechanisms of initiation, and roles of activators and coactivators
Abstract
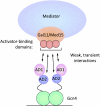
Here we review recent advances in understanding the regulation of mRNA synthesis in Saccharomyces cerevisiae. Many fundamental gene regulatory mechanisms have been conserved in all eukaryotes, and budding yeast has been at the forefront in the discovery and dissection of these conserved mechanisms. Topics covered include upstream activation sequence and promoter structure, transcription factor classification, and examples of regulated transcription factor activity. We also examine advances in understanding the RNA polymerase II transcription machinery, conserved coactivator complexes, transcription activation domains, and the cooperation of these factors in gene regulatory mechanisms.
Figures




References
-
- Agricola E., Verdone L., Xella B., Di Mauro E., Caserta M., 2004. Common chromatin architecture, common chromatin remodeling, and common transcription kinetics of Adr1-dependent genes in Saccharomyces cerevisiae. Biochemistry 43: 8878–8884 - PubMed
-
- Agricola E., Verdone L., Di Mauro E., Caserta M., 2006. H4 acetylation does not replace H3 acetylation in chromatin remodelling and transcription activation of Adr1-dependent genes. Mol. Microbiol. 62: 1433–1446 - PubMed
-
- Albert I., Mavrich T. N., Tomsho L. P., Qi J., Zanton S. J., et al. , 2007. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature 446: 572–576 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

