Emergence and evolution of the glycoprotein hormone and neurotrophin gene families in vertebrates
- PMID: 22085792
- PMCID: PMC3280201
- DOI: 10.1186/1471-2148-11-332
Emergence and evolution of the glycoprotein hormone and neurotrophin gene families in vertebrates
Abstract
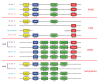
Background: The three vertebrate pituitary glycoprotein hormones (GPH) are heterodimers of a common α and a specific β subunit. In human, they are located on different chromosomes but in a similar genomic environment. We took advantage of the availability of genomic and EST data from two cartilaginous fish species as well as from two lamprey species to identify their repertoire of neurotrophin, lin7 and KCNA gene family members which are in the close environment of gphβ. Gphα and gphβ are absent outside vertebrates but are related to two genes present in both protostomes and deuterostomes that were named gpa2 and gpb5. Genomic organization and functional characteristics of their protein products suggested that gphα and gphβ might have been generated concomitantly by a duplication of gpa2 and gpb5 just prior to the radiation of vertebrates. To have a better insight into this process we used new genomic resources and tools to characterize the ancestral environment before the duplication occurred.
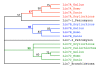
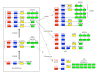
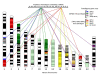
Results: An almost similar repertoire of genes was characterized in cartilaginous fishes as in tetrapods. Data in lampreys are either incomplete or the result of specific duplications and/or deletions but a scenario for the evolution of this genomic environment in vertebrates could be proposed. A number of genes were identified in the amphioxus genome that helped in reconstructing the ancestral environment of gpa2 and gpb5 and in describing the evolution of this environment in vertebrates.
Conclusion: Our model suggests that vertebrate gphα and gphβ were generated by a specific local duplication of the ancestral forms of gpa2 and gpb5, followed by a translocation of gphβ to a new environment whereas gphα was retained in the gpa2-gpb5 locus. The two rounds of whole genome duplication that occurred early in the evolution of vertebrates generated four paralogues of each gene but secondary gene losses or lineage specific duplications together with genomic rearrangements have resulted in the present organization of these genes, which differs between vertebrate lineages.
Figures




References
-
- Quérat B, Sellouk A, Salmon C. Phylogenetic analysis of the vertebrate glycoprotein hormone family including new sequences of sturgeon Acipenser baeri, β-subunits of the two gonadotropins and the thyroid-stimulating hormone. Biol Reprod. 2000;63:222–228. doi: 10.1095/biolreprod63.1.222. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

