Optimal functional levels of activation-induced deaminase specifically require the Hsp40 DnaJa1
- PMID: 22085931
- PMCID: PMC3273393
- DOI: 10.1038/emboj.2011.417
Optimal functional levels of activation-induced deaminase specifically require the Hsp40 DnaJa1
Abstract
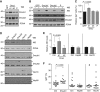
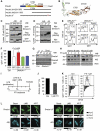
The enzyme activation-induced deaminase (AID) deaminates deoxycytidine at the immunoglobulin genes, thereby initiating antibody affinity maturation and isotype class switching during immune responses. In contrast, off-target DNA damage caused by AID is oncogenic. Central to balancing immunity and cancer is AID regulation, including the mechanisms determining AID protein levels. We describe a specific functional interaction between AID and the Hsp40 DnaJa1, which provides insight into the function of both proteins. Although both major cytoplasmic type I Hsp40s, DnaJa1 and DnaJa2, are induced upon B-cell activation and interact with AID in vitro, only DnaJa1 overexpression increases AID levels and biological activity in cell lines. Conversely, DnaJa1, but not DnaJa2, depletion reduces AID levels, stability and isotype switching. In vivo, DnaJa1-deficient mice display compromised response to immunization, AID protein and isotype switching levels being reduced by half. Moreover, DnaJa1 farnesylation is required to maintain, and farnesyltransferase inhibition reduces, AID protein levels in B cells. Thus, DnaJa1 is a limiting factor that plays a non-redundant role in the functional stabilization of AID.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures




Similar articles
-
Inhibition of mutant Kras and p53-driven pancreatic carcinogenesis by atorvastatin: Mainly via targeting of the farnesylated DNAJA1 in chaperoning mutant p53.Mol Carcinog. 2019 Nov;58(11):2052-2064. doi: 10.1002/mc.23097. Epub 2019 Aug 9. Mol Carcinog. 2019. PMID: 31397499 Free PMC article.
-
Heat shock protein DNAJA1 stabilizes PIWI proteins to support regeneration and homeostasis of planarian Schmidtea mediterranea.J Biol Chem. 2019 Jun 21;294(25):9873-9887. doi: 10.1074/jbc.RA118.004445. Epub 2019 May 10. J Biol Chem. 2019. PMID: 31076507 Free PMC article.
-
Identification of a druggable protein-protein interaction site between mutant p53 and its stabilizing chaperone DNAJA1.J Biol Chem. 2021 Jan-Jun;296:100098. doi: 10.1074/jbc.RA120.014749. Epub 2020 Nov 21. J Biol Chem. 2021. PMID: 33208462 Free PMC article.
-
AID in Antibody Diversification: There and Back Again.Trends Immunol. 2020 Jul;41(7):586-600. doi: 10.1016/j.it.2020.04.009. Epub 2020 Apr 27. Trends Immunol. 2020. PMID: 32434680 Free PMC article. Review.
-
Activation-induced deaminase in B lymphocyte maturation and beyond.Biomed J. 2013 Nov-Dec;36(6):259-68. doi: 10.4103/2319-4170.113191. Biomed J. 2013. PMID: 24385067 Review.
Cited by
-
AID/APOBEC deaminases and cancer.Oncoscience. 2015 Apr 28;2(4):320-33. doi: 10.18632/oncoscience.155. eCollection 2015. Oncoscience. 2015. PMID: 26097867 Free PMC article. Review.
-
A cytosolic chaperone complex controls folding and degradation of type III CD38.J Biol Chem. 2019 Mar 15;294(11):4247-4258. doi: 10.1074/jbc.RA118.005844. Epub 2019 Jan 22. J Biol Chem. 2019. PMID: 30670591 Free PMC article.
-
The Role of APOBECs in Viral Replication.Microorganisms. 2020 Nov 30;8(12):1899. doi: 10.3390/microorganisms8121899. Microorganisms. 2020. PMID: 33266042 Free PMC article. Review.
-
Selective Binding of HSC70 and its Co-Chaperones to Structural Hotspots on CFTR.Sci Rep. 2020 Mar 6;10(1):4176. doi: 10.1038/s41598-020-61107-x. Sci Rep. 2020. PMID: 32144307 Free PMC article.
-
Landscape of transcription and expression regulated by DNA methylation related to age of donor and cell passage in adipose-derived mesenchymal stem cells.Aging (Albany NY). 2020 Oct 31;12(21):21186-21201. doi: 10.18632/aging.103809. Epub 2020 Oct 31. Aging (Albany NY). 2020. PMID: 33130636 Free PMC article.
References
-
- Arakawa H, Hauschild J, Buerstedde J-M (2002) Requirement of the activation-induced deaminase (AID) gene for immunoglobulin gene conversion. Science 295: 1301–1306 - PubMed
-
- Bardwell JC, Tilly K, Craig E, King J, Zylicz M, Georgopoulos C (1986) The nucleotide sequence of the Escherichia coli K12 dnaJ+ gene. A gene that encodes a heat shock protein. J Biol Chem 261: 1782–1785 - PubMed
-
- Brar SS, Watson M, Diaz M (2004) Activation-induced cytosine deaminase (AID) is actively exported out of the nucleus but retained by the induction of DNA breaks. J Biol Chem 279: 26395–26401 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

