A master CLOCK hard at work brings rhythm to the transcriptome
- PMID: 22085960
- PMCID: PMC3222898
- DOI: 10.1101/gad.180984.111
A master CLOCK hard at work brings rhythm to the transcriptome
Abstract
In this issue of Genes & Development, Abruzzi et al. (pp. 2374-2386) use chromatin immunoprecipitation (ChIP) tiling array assays (ChIP-chip) to show that physical interactions between circadian (≅24-h) clock machineries and genomes are more widespread than previously thought and provide novel insights into how clocks drive daily rhythms in global gene expression.
Figures
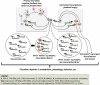
Comment on
-
Drosophila CLOCK target gene characterization: implications for circadian tissue-specific gene expression.Genes Dev. 2011 Nov 15;25(22):2374-86. doi: 10.1101/gad.178079.111. Genes Dev. 2011. PMID: 22085964 Free PMC article.
References
-
- Allada R, Emery P, Takahashi JS, Rosbash M 2001. Stopping time: The genetics of fly and mouse circadian clocks. Annu Rev Neurosci 24: 1091–1119 - PubMed
-
- Bae K, Edery I 2006. Regulating a circadian clock's period, phase and amplitude by phosphorylation: Insights from Drosophila. J Biochem 140: 609–617 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
