Competition for cofactor-dependent DNA binding underlies Hox phenotypic suppression
- PMID: 22085961
- PMCID: PMC3222899
- DOI: 10.1101/gad.175539.111
Competition for cofactor-dependent DNA binding underlies Hox phenotypic suppression
Abstract
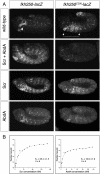
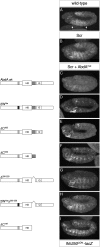
Hox transcription factors exhibit an evolutionarily conserved functional hierarchy, termed phenotypic suppression, in which the activity of posterior Hox proteins dominates over more anterior Hox proteins. Using directly regulated Hox targeted reporter genes in Drosophila, we show that posterior Hox proteins suppress the activities of anterior ones by competing for cofactor-dependent DNA binding. Furthermore, we map a motif in the posterior Hox protein Abdominal-A (AbdA) that is required for phenotypic suppression and facilitates cooperative DNA binding with the Hox cofactor Extradenticle (Exd). Together, these results suggest that Hox-specific motifs endow posterior Hox proteins with the ability to dominate over more anterior ones via a cofactor-dependent DNA-binding mechanism.
Figures
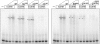

Similar articles
-
Hox repression of a target gene: extradenticle-independent, additive action through multiple monomer binding sites.Development. 2002 Jul;129(13):3115-26. doi: 10.1242/dev.129.13.3115. Development. 2002. PMID: 12070087
-
A unique Extradenticle recruitment mode in the Drosophila Hox protein Ultrabithorax.Proc Natl Acad Sci U S A. 2007 Oct 23;104(43):16946-51. doi: 10.1073/pnas.0705832104. Epub 2007 Oct 17. Proc Natl Acad Sci U S A. 2007. PMID: 17942685 Free PMC article.
-
Regulation of Hox target genes by a DNA bound Homothorax/Hox/Extradenticle complex.Development. 1999 Nov;126(22):5137-48. doi: 10.1242/dev.126.22.5137. Development. 1999. PMID: 10529430
-
Cellular analysis of newly identified Hox downstream genes in Drosophila.Eur J Cell Biol. 2010 Feb-Mar;89(2-3):273-8. doi: 10.1016/j.ejcb.2009.11.012. Epub 2009 Dec 16. Eur J Cell Biol. 2010. PMID: 20018403 Review.
-
Hox cofactors in vertebrate development.Dev Biol. 2006 Mar 15;291(2):193-206. doi: 10.1016/j.ydbio.2005.10.032. Epub 2006 Mar 3. Dev Biol. 2006. PMID: 16515781 Review.
Cited by
-
Using competition assays to quantitatively model cooperative binding by transcription factors and other ligands.Biochim Biophys Acta Gen Subj. 2017 Nov;1861(11 Pt A):2789-2801. doi: 10.1016/j.bbagen.2017.07.024. Epub 2017 Aug 1. Biochim Biophys Acta Gen Subj. 2017. PMID: 28774855 Free PMC article.
-
Hox Genes Promote Neuronal Subtype Diversification through Posterior Induction in Caenorhabditis elegans.Neuron. 2015 Nov 4;88(3):514-27. doi: 10.1016/j.neuron.2015.09.049. Neuron. 2015. PMID: 26539892 Free PMC article.
-
Diversification and Functional Evolution of HOX Proteins.Front Cell Dev Biol. 2022 May 13;10:798812. doi: 10.3389/fcell.2022.798812. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35646905 Free PMC article. Review.
-
Hox proteins display a common and ancestral ability to diversify their interaction mode with the PBC class cofactors.PLoS Biol. 2012;10(6):e1001351. doi: 10.1371/journal.pbio.1001351. Epub 2012 Jun 26. PLoS Biol. 2012. PMID: 22745600 Free PMC article.
-
Role of abd-A and Abd-B in development of abdominal epithelia breaks posterior prevalence rule.PLoS Genet. 2014 Oct 23;10(10):e1004717. doi: 10.1371/journal.pgen.1004717. eCollection 2014 Oct. PLoS Genet. 2014. PMID: 25340649 Free PMC article.
References
-
- Abu-Shaar M, Mann RS 1998. Generation of multiple antagonistic domains along the proximodistal axis during Drosophila leg development. Development 125: 3821–3830 - PubMed
-
- Brand AH, Perrimon N 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
