Tumor-specific retargeting of an oncogenic transcription factor chimera results in dysregulation of chromatin and transcription
- PMID: 22086061
- PMCID: PMC3266033
- DOI: 10.1101/gr.125666.111
Tumor-specific retargeting of an oncogenic transcription factor chimera results in dysregulation of chromatin and transcription
Abstract
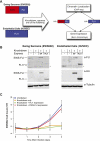
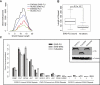
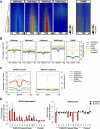
Chromosomal translocations involving transcription factor genes have been identified in an increasingly wide range of cancers. Some translocations can create a protein "chimera" that is composed of parts from different proteins. How such chimeras cause cancer, and why they cause cancer in some cell types but not others, is not understood. One such chimera is EWS-FLI, the most frequently occurring translocation in Ewing Sarcoma, a malignant bone and soft tissue tumor of children and young adults. Using EWS-FLI and its parental transcription factor, FLI1, we created a unique experimental system to address questions regarding the genomic mechanisms by which chimeric transcription factors cause cancer. We found that in tumor cells, EWS-FLI targets regions of the genome distinct from FLI1, despite identical DNA-binding domains. In primary endothelial cells, however, EWS-FLI and FLI1 demonstrate similar targeting. To understand this mistargeting, we examined chromatin organization. Regions targeted by EWS-FLI are normally repressed and nucleosomal in primary endothelial cells. In tumor cells, however, bound regions are nucleosome depleted and harbor the chromatin signature of enhancers. We next demonstrated that through chimerism, EWS-FLI acquired the ability to alter chromatin. Expression of EWS-FLI results in nucleosome depletion at targeted sites, whereas silencing of EWS-FLI in tumor cells restored nucleosome occupancy. Thus, the EWS-FLI chimera acquired chromatin-altering activity, leading to mistargeting, chromatin disruption, and ultimately, transcriptional dysregulation.
Figures



References
-
- Bartel FO, Higuchi T, Spyropoulos DD 2000. Mouse models in the study of the Ets family of transcription factors. Oncogene 19: 6443–6454 - PubMed
-
- Bassuk AG, Leiden JM 1995. A direct physical association between ETS and AP-1 transcription factors in normal human T cells. Immunity 3: 223–237 - PubMed
-
- Benjamin LE, Fredericks WJ, Barr FG, Rauscher FJ 3rd 1996. Fusion of the EWS1 and WT1 genes as a result of the t(11;22)(p13;q12) translocation in desmoplastic small round cell tumors. Med Pediatr Oncol 27: 434–439 - PubMed
-
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
