The origin and evolution of vertebrate sex chromosomes and dosage compensation
- PMID: 22086077
- PMCID: PMC3238118
- DOI: 10.1038/hdy.2011.106
The origin and evolution of vertebrate sex chromosomes and dosage compensation
Abstract
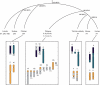
In mammals, birds, snakes and many lizards and fish, sex is determined genetically (either male XY heterogamy or female ZW heterogamy), whereas in alligators, and in many reptiles and turtles, the temperature at which eggs are incubated determines sex. Evidently, different sex-determining systems (and sex chromosome pairs) have evolved independently in different vertebrate lineages. Homology shared by Xs and Ys (and Zs and Ws) within species demonstrates that differentiated sex chromosomes were once homologous, and that the sex-specific non-recombining Y (or W) was progressively degraded. Consequently, genes are left in single copy in the heterogametic sex, which results in an imbalance of the dosage of genes on the sex chromosomes between the sexes, and also relative to the autosomes. Dosage compensation has evolved in diverse species to compensate for these dose differences, with the stringency of compensation apparently differing greatly between lineages, perhaps reflecting the concentration of genes on the original autosome pair that required dosage compensation. We discuss the organization and evolution of amniote sex chromosomes, and hypothesize that dosage insensitivity might predispose an autosome to evolving function as a sex chromosome.
Figures

References
-
- Barber JC. Terminal 3p deletions: phenotypic variability, chromosomal non-penetrance, or gene modification. Am J Med Genet A. 2008;146A:1899–1901. - PubMed
-
- Baverstock PR, Adams M, Polkinghorne RW, Gelder M. A sex-linked enzyme in birds—Z-chromosome conservation but no dosage compensation. Nature. 1982;296:763–766. - PubMed
-
- Bianchi NO, Becak W, De Bianchi MS, Becak ML, Rabello MN. Chromosome replication in four species of snakes. Chromosoma. 1969;26:188–200. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

