MetaCrop 2.0: managing and exploring information about crop plant metabolism
- PMID: 22086948
- PMCID: PMC3245004
- DOI: 10.1093/nar/gkr1004
MetaCrop 2.0: managing and exploring information about crop plant metabolism
Abstract
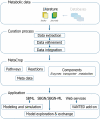
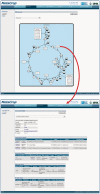
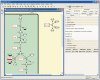
MetaCrop is a manually curated repository of high-quality data about plant metabolism, providing different levels of detail from overview maps of primary metabolism to kinetic data of enzymes. It contains information about seven major crop plants with high agronomical importance and two model plants. MetaCrop is intended to support research aimed at the improvement of crops for both nutrition and industrial use. It can be accessed via web, web services and an add-on to the Vanted software. Here, we present several novel developments of the MetaCrop system and the extended database content. MetaCrop is now available in version 2.0 at http://metacrop.ipk-gatersleben.de.
Figures


References
-
- Le Novère N, Hucka M, Mi H, Moodie S, Schreiber F, Sorokin A, Demir E, Wegner K, Aladjem MI, Wimalaratne SM, et al. The Systems Biology Graphical Notation. Nat. Biotech. 2009;27:735–741. - PubMed
-
- Hucka M, Finney A, Sauro HM, Bolouri H, Doyle JC, Kitano H, Arkin AP, Bornstein BJ, Bray D, Cornish-Bowden A, et al. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19:524–531. - PubMed

