doRiNA: a database of RNA interactions in post-transcriptional regulation
- PMID: 22086949
- PMCID: PMC3245013
- DOI: 10.1093/nar/gkr1007
doRiNA: a database of RNA interactions in post-transcriptional regulation
Abstract
In animals, RNA binding proteins (RBPs) and microRNAs (miRNAs) post-transcriptionally regulate the expression of virtually all genes by binding to RNA. Recent advances in experimental and computational methods facilitate transcriptome-wide mapping of these interactions. It is thought that the combinatorial action of RBPs and miRNAs on target mRNAs form a post-transcriptional regulatory code. We provide a database that supports the quest for deciphering this regulatory code. Within doRiNA, we are systematically curating, storing and integrating binding site data for RBPs and miRNAs. Users are free to take a target (mRNA) or regulator (RBP and/or miRNA) centric view on the data. We have implemented a database framework with short query response times for complex searches (e.g. asking for all targets of a particular combination of regulators). All search results can be browsed, inspected and analyzed in conjunction with a huge selection of other genome-wide data, because our database is directly linked to a local copy of the UCSC genome browser. At the time of writing, doRiNA encompasses RBP data for the human, mouse and worm genomes. For computational miRNA target site predictions, we provide an update of PicTar predictions.
Figures

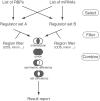
References
-
- Rajewsky N. MicroRNAs and the Operon paper. J. Mol. Biol. 2011;409:70–75. - PubMed
-
- Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010;11:597–610. - PubMed
-
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 2008;9:102–114. - PubMed
-
- Krek A, Grün D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, et al. Combinatorial microRNA target predictions. Net. Genet. 2005;37:495–500. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

