Serum microRNA expression profile distinguishes enterovirus 71 and coxsackievirus 16 infections in patients with hand-foot-and-mouth disease
- PMID: 22087245
- PMCID: PMC3210764
- DOI: 10.1371/journal.pone.0027071
Serum microRNA expression profile distinguishes enterovirus 71 and coxsackievirus 16 infections in patients with hand-foot-and-mouth disease
Abstract
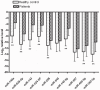
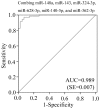
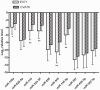
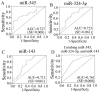
Altered circulating microRNA (miRNA) profiles have been noted in patients with microbial infections. We compared host serum miRNA levels in patients with hand-foot-and-mouth disease (HFMD) caused by enterovirus 71 (EV71) and coxsackievirus 16 (CVA16) as well as in other microbial infections and in healthy individuals. Among 664 different miRNAs analyzed using a miRNA array, 102 were up-regulated and 26 were down-regulated in sera of patients with enteroviral infections. Expression levels of ten candidate miRNAs were further evaluated by quantitative real-time PCR assays. A receiver operating characteristic (ROC) curve analysis revealed that six miRNAs (miR-148a, miR-143, miR-324-3p, miR-628-3p, miR-140-5p, and miR-362-3p) were able to discriminate patients with enterovirus infections from healthy controls with area under curve (AUC) values ranged from 0.828 to 0.934. The combined six miRNA using multiple logistic regression analysis provided not only a sensitivity of 97.1% and a specificity of 92.7% but also a unique profile that differentiated enterovirial infections from other microbial infections. Expression levels of five miRNAs (miR-148a, miR-143, miR-324-3p, miR-545, and miR-140-5p) were significantly increased in patients with CVA16 versus those with EV71 (p<0.05). Combination of miR-545, miR-324-3p, and miR-143 possessed a moderate ability to discrimination between CVA16 and EV71 with an AUC value of 0.761. These data indicate that sera from patients with different subtypes of enteroviral infection express unique miRNA profiles. Serum miRNA expression profiles may provide supplemental biomarkers for diagnosing and subtyping enteroviral HFMD infections.
Conflict of interest statement
Figures
Similar articles
-
MicroRNA expression profile in exosome discriminates extremely severe infections from mild infections for hand, foot and mouth disease.BMC Infect Dis. 2014 Sep 17;14:506. doi: 10.1186/1471-2334-14-506. BMC Infect Dis. 2014. PMID: 25231540 Free PMC article.
-
Elevated circulating miR-494 in plasma of children with enterovirus 71 induced hand, foot, and mouth disease and its potential diagnostic value.Acta Virol. 2020;64(3):338-343. doi: 10.4149/av_2020_311. Acta Virol. 2020. PMID: 32985212
-
Comparative analysis of putative novel microRNA expression profiles induced by enterovirus 71 and coxsackievirus A16 infections in human umbilical vein endothelial cells using high-throughput sequencing.Infect Genet Evol. 2019 Sep;73:401-410. doi: 10.1016/j.meegid.2019.06.007. Epub 2019 Jun 5. Infect Genet Evol. 2019. PMID: 31176031
-
Coxsackievirus A6: a new emerging pathogen causing hand, foot and mouth disease outbreaks worldwide.Expert Rev Anti Infect Ther. 2015;13(9):1061-71. doi: 10.1586/14787210.2015.1058156. Epub 2015 Jun 25. Expert Rev Anti Infect Ther. 2015. PMID: 26112307 Review.
-
Coxsackievirus A6 associated hand, foot and mouth disease in adults: clinical presentation and review of the literature.J Clin Virol. 2014 Aug;60(4):381-6. doi: 10.1016/j.jcv.2014.04.023. Epub 2014 May 9. J Clin Virol. 2014. PMID: 24932735 Review.
Cited by
-
MicroRNA 876-5p modulates EV-A71 replication through downregulation of host antiviral factors.Virol J. 2020 Feb 5;17(1):21. doi: 10.1186/s12985-020-1284-8. Virol J. 2020. PMID: 32024541 Free PMC article.
-
Dysregulated microRNA expression in serum of non-vaccinated children with varicella.Viruses. 2014 Apr 22;6(4):1823-36. doi: 10.3390/v6041823. Viruses. 2014. PMID: 24759212 Free PMC article.
-
miRNome analysis reveals mir-155-5p as a protective factor to dengue infection in a resistant Thai cohort.Med Microbiol Immunol. 2025 Feb 20;214(1):13. doi: 10.1007/s00430-025-00821-7. Med Microbiol Immunol. 2025. PMID: 39976655 Free PMC article.
-
Innate Immunity Evasion by Enteroviruses: Insights into Virus-Host Interaction.Viruses. 2016 Jan 15;8(1):22. doi: 10.3390/v8010022. Viruses. 2016. PMID: 26784219 Free PMC article. Review.
-
Circulating microRNAs as Potential Biomarkers of Infectious Disease.Front Immunol. 2017 Feb 16;8:118. doi: 10.3389/fimmu.2017.00118. eCollection 2017. Front Immunol. 2017. PMID: 28261201 Free PMC article. Review.
References
-
- Ding NZ, Wang XM, Sun SW, Song Q, Li SN, et al. Appearance of mosaic enterovirus 71 in the 2008 outbreak of China. Virus Res. 2009;145:157–161. - PubMed
-
- The Chinese Center for Disease Control and Prevention. The Office of the World Health Organization in China. Report on the Hand, Foot and Mouth Disease Outbreak in Fuyang City, Anhui Province and the Prevention and Control in China. 2008. Available: http://www.wpro.who.int/NR/rdonlyres/591D6A7B-FB15-4E94-A1E9-1D3381847D6....
-
- Shih SR, Stollar V, Lin JY, Chang SC, Chen GW, et al. Identification of genes involved in the host response to enterovirus 71 infection. J Neurovirol. 2004;10:293–304. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

