Dependence of avidity on linker length for a bivalent ligand-bivalent receptor model system
- PMID: 22088143
- PMCID: PMC3272676
- DOI: 10.1021/ja2073033
Dependence of avidity on linker length for a bivalent ligand-bivalent receptor model system
Abstract
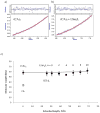
This paper describes a synthetic dimer of carbonic anhydrase, and a series of bivalent sulfonamide ligands with different lengths (25 to 69 Å between the ends of the fully extended ligands), as a model system to use in examining the binding of bivalent antibodies to antigens. Assays based on analytical ultracentrifugation and fluorescence binding indicate that this system forms cyclic, noncovalent complexes with a stoichiometry of one bivalent ligand to one dimer. This dimer binds the series of bivalent ligands with low picomolar avidities (K(d)(avidity) = 3-40 pM). A structurally analogous monovalent ligand binds to one active site of the dimer with K(d)(mono) = 16 nM. The bivalent association is thus significantly stronger (K(d)(mono)/K(d)(avidity) ranging from ~500 to 5000 unitless) than the monovalent association. We infer from these results, and by comparison of these results to previous studies, that bivalency in antibodies can lead to associations much tighter than monovalent associations (although the observed bivalent association is much weaker than predicted from the simplest level of theory: predicted K(d)(avidity) of ~0.002 pM and K(d)(mono)/K(d)(avidity) ~ 8 × 10(6) unitless).
© 2011 American Chemical Society
Figures
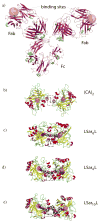







References
-
- Murphy K, Travers P, Walport M. Janeway’s Immunobiology. 7. Garland Science, Taylor & Francis Group, LLC; New York and London: 2008.
-
- Houk KN, Leach AG, Kim SP, Zhang XY. Angew Chem, Int Ed Eng. 2003;42:4872. - PubMed
-
- Gestwicki JE, Cairo CW, Strong LE, Oetjen KA, Kiessling LL. J Am Chem Soc. 2002;124:14922. - PubMed
-
- Mourez M, Collier RJ. Use of Phage Display and Polyvalency to Design Inhibitors of Protein-Protein Interactions. Vol. 261 Humana Press Inc; Totowa, NJ: 2004. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

