Understanding biomolecular motion, recognition, and allostery by use of conformational ensembles
- PMID: 22089251
- PMCID: PMC3222826
- DOI: 10.1007/s00249-011-0754-8
Understanding biomolecular motion, recognition, and allostery by use of conformational ensembles
Abstract
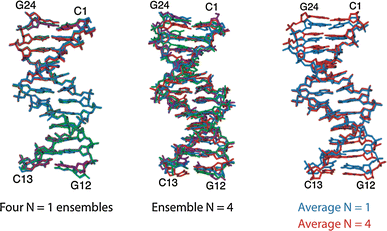
We review the role conformational ensembles can play in the analysis of biomolecular dynamics, molecular recognition, and allostery. We introduce currently available methods for generating ensembles of biomolecules and illustrate their application with relevant examples from the literature. We show how, for binding, conformational ensembles provide a way of distinguishing the competing models of induced fit and conformational selection. For allostery we review the classic models and show how conformational ensembles can play a role in unravelling the intricate pathways of communication that enable allostery to occur. Finally, we discuss the limitations of conformational ensembles and highlight some potential applications for the future.
Figures









References
-
- Allerhand A, Doddrell D, Glushko V, Cochran DW, Wenkert E, Lawson PJ, Gurd FR. Conformation and segmental motion of native and denatured ribonuclease A in solution. Application of natural-abundance carbon-13 partially relaxed Fourier transform nuclear magnetic resonance. J Am Chem Soc. 1971;93:544–546. doi: 10.1021/ja00731a053. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

