Monoclonal antibody-based antigenic mapping of norovirus GII.4-2002
- PMID: 22090098
- PMCID: PMC3255811
- DOI: 10.1128/JVI.06200-11
Monoclonal antibody-based antigenic mapping of norovirus GII.4-2002
Abstract
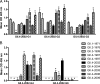
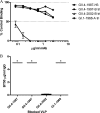
Noroviruses are the primary cause of epidemic gastroenteritis in humans, and GII.4 strains cause ∼80% of the overall disease burden. Surrogate neutralization assays using sera and mouse monoclonal antibodies (MAbs) suggest that antigenic variation maintains GII.4 persistence in the face of herd immunity, as the emergence of new pandemic strains is accompanied by newly evolved neutralization epitopes. To potentially identify specific blockade epitopes that are likely neutralizing and evolving between pandemic strains, mice were hyperimmunized with GII.4-2002 virus-like particles (VLPs) and the resulting MAbs were characterized by biochemical and immunologic assays. All of the MAbs but one recognized GII.4 VLPs representing strains circulating from 1987 to 2009. One MAb weakly recognized GII.4-1987 and -1997 while strongly interacting with 2002 VLPs. This antibody was highly selective and effective at blocking only GII.4-2002-ligand binding. Using bioinformatic analyses, we predicted an evolving GII.4 surface epitope composed of amino acids 407, 412, and 413 and subsequently built mutant VLPs to test the impact of the epitope on MAb binding and blockade potential. Replacement of the 2002 epitope with the epitopes found in 1987 or 2006 strains either reduced or ablated enzyme immunoassay recognition by the GII.4-2002-specific blockade MAb. These data identify a novel, evolving blockade epitope that may be associated with protective immunity, providing further support for the hypotheses that GII.4 norovirus evolution results in antigenic variation that allows the virus to escape from protective herd immunity, resulting in new epidemic strains.
Figures







References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

