Significant association of multiple human cytomegalovirus genomic Loci with glioblastoma multiforme samples
- PMID: 22090104
- PMCID: PMC3255835
- DOI: 10.1128/JVI.06097-11
Significant association of multiple human cytomegalovirus genomic Loci with glioblastoma multiforme samples
Abstract
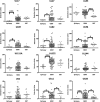
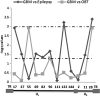
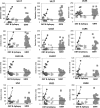
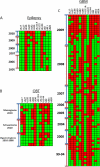
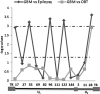
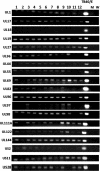
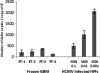
Viruses are appreciated as etiological agents of certain human tumors, but the number of different cancer types induced or exacerbated by viral infections is unknown. Glioblastoma multiforme (GBM)/astrocytoma grade IV is a malignant and lethal brain cancer of unknown origin. Over the past decade, several studies have searched for the presence of a prominent herpesvirus, human cytomegalovirus (HCMV), in GBM samples. While some have detected HCMV DNA, RNA, and proteins in GBM tissues, others have not. Therefore, any purported association of HCMV with GBM remains controversial. In most of the previous studies, only one or a select few viral targets were analyzed. Thus, it remains unclear the extent to which the entire viral genome was present when detected. Here we report the results of a survey of GBM specimens for as many as 20 different regions of the HCMV genome. Our findings indicate that multiple HCMV loci are statistically more likely to be found in GBM samples than in other brain tumors or epileptic brain specimens and that the viral genome was more often detected in frozen samples than in paraffin-embedded archival tissue samples. Finally, our experimental results indicate that cellular genomes substantially outnumber viral genomes in HCMV-positive GBM specimens, likely indicating that only a minority of the cells found in such samples harbor viral DNA. These data argue for the association of HCMV with GBM, defining the virus as oncoaccessory. Furthermore, they imply that, were HCMV to enhance the growth or survival of a tumor (i.e., if it is oncomodulatory), it would likely do so through mechanisms distinct from classic tumor viruses that express transforming viral oncoproteins in the overwhelming majority of tumor cells.
Figures

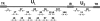


References
-
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate - a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57:289–300
-
- Britt W. 2008. Manifestations of human cytomegalovirus infection: proposed mechanisms of acute and chronic disease. Curr. Top. Microbiol. Immunol. 325:417–470 - PubMed
-
- Brune W. 2011. Inhibition of programmed cell death by cytomegaloviruses. Virus Res. 157:144–150 - PubMed
-
- Cobbs CS, et al. 2002. Human cytomegalovirus infection and expression in human malignant glioma. Cancer Res. 62:3347–3350 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

