Migration of Norway rats resulted in the worldwide distribution of Seoul hantavirus today
- PMID: 22090114
- PMCID: PMC3255798
- DOI: 10.1128/JVI.00725-11
Migration of Norway rats resulted in the worldwide distribution of Seoul hantavirus today
Abstract
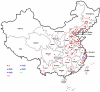
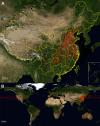
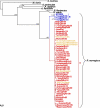
Despite the worldwide distribution, most of the known Seoul viruses (SEOV) are closely related to each other. In this study, the M and the S segment sequences of SEOV were recovered from 130 lung tissue samples (mostly of Norway rats) and from six patient serum samples by reverse transcription-PCR. Genetic analysis revealed that all sequences belong to SEOV and represent 136 novel strains. Phylogenetic analysis of all available M and S segment sequences of SEOV, including 136 novel Chinese strains, revealed four distinct groups. All non-Chinese SEOV strains and most of the Chinese variants fell into the phylogroup A, while the Chinese strains originating from mountainous areas clustered into three other distinct groups (B, C, and D). We estimated that phylogroup A viruses may have arisen only within the last several centuries. All non-Chinese variants appeared to be directly originated from China. Thus, phylogroup A viruses distributed worldwide may share a recent ancestor, whereas SEOV seems to be as diversified genetically as other hantaviruses. In addition, all available mitochondrial DNA (mtDNA) sequences of Norway rats, including our 44 newly recovered mtDNA sequences, were divided into two phylogenetic groups. The first group, which is associated with the group A SEOV variants, included most of rats from China and also all non-Chinese rats, while the second group consisted of a few rats originating only from mountain areas in China. We hypothesize that an ancestor of phylogroup A SEOV variants was first exported from China to Europe and then spread through the New World following the migration of Norway rats.
Figures


References
-
- . 2004. History of the Norway rat (Rattus norvegicus). http://www.ratbehavior.org/images/history.htm Accessed 18 November 2011
-
- Barnett SA. 2002. The story of rats: their impact on us, and our impact on them, p 17–18 Allen and Unwin, Crows Nest, Australia
-
- Chen H, et al. 2005. Avian flu: H5N1 virus outbreak in migratory waterfowl. Nature 436:191–192 - PubMed
-
- Chen HX, Luo CW. 2001. Hemorrhagic fever with renal syndrome: studies and the surveillance and application of vaccine. Hong Kong Medical Publisher, Hong Kong
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

