YPR139c/LOA1 encodes a novel lysophosphatidic acid acyltransferase associated with lipid droplets and involved in TAG homeostasis
- PMID: 22090344
- PMCID: PMC3258169
- DOI: 10.1091/mbc.E11-07-0650
YPR139c/LOA1 encodes a novel lysophosphatidic acid acyltransferase associated with lipid droplets and involved in TAG homeostasis
Abstract
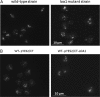
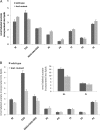
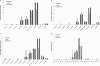
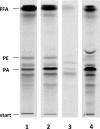
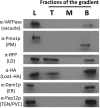
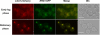
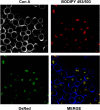
For many years, lipid droplets (LDs) were considered to be an inert store of lipids. However, recent data showed that LDs are dynamic organelles playing an important role in storage and mobilization of neutral lipids. In this paper, we report the characterization of LOA1 (alias VPS66, alias YPR139c), a yeast member of the glycerolipid acyltransferase family. LOA1 mutants show abnormalities in LD morphology. As previously reported, cells lacking LOA1 contain more LDs. Conversely, we showed that overexpression results in fewer LDs. We then compared the lipidome of loa1Δ mutant and wild-type strains. Steady-state metabolic labeling of loa1Δ revealed a significant reduction in triacylglycerol content, while phospholipid (PL) composition remained unchanged. Interestingly, lipidomic analysis indicates that both PLs and glycerolipids are qualitatively affected by the mutation, suggesting that Loa1p is a lysophosphatidic acid acyltransferase (LPA AT) with a preference for oleoyl-CoA. This hypothesis was tested by in vitro assays using both membranes of Escherichia coli cells expressing LOA1 and purified proteins as enzyme sources. Our results from purification of subcellular compartments and proteomic studies show that Loa1p is associated with LD and active in this compartment. Loa1p is therefore a novel LPA AT and plays a role in LD formation.
Figures




References
-
- Beaudoin F, Wilkinson BM, Stirling CJ, Napier JA. In vivo targeting of a sunflower oil body protein in yeast secretory (sec) mutants. Plant J. 2000;23:159–70. - PubMed
-
- Benghezal M, Roubaty C, Veepuri V, Knudsen J, Conzelmann A. SLC1 and SLC4 encode partially redundant acyl-coenzyme A 1-acylglycerol-3-phosphate O-acyltransferases of budding yeast. J Biol Chem. 2007;282:30845–30855. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

