Interlocus gene conversion events introduce deleterious mutations into at least 1% of human genes associated with inherited disease
- PMID: 22090377
- PMCID: PMC3290778
- DOI: 10.1101/gr.127738.111
Interlocus gene conversion events introduce deleterious mutations into at least 1% of human genes associated with inherited disease
Abstract
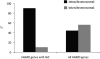
Establishing the molecular basis of DNA mutations that cause inherited disease is of fundamental importance to understanding the origin, nature, and clinical sequelae of genetic disorders in humans. The majority of disease-associated mutations constitute single-base substitutions and short deletions and/or insertions resulting from DNA replication errors and the repair of damaged bases. However, pathological mutations can also be introduced by nonreciprocal recombination events between paralogous sequences, a phenomenon known as interlocus gene conversion (IGC). IGC events have thus far been linked to pathology in more than 20 human genes. However, the large number of duplicated gene sequences in the human genome implies that many more disease-associated mutations could originate via IGC. Here, we have used a genome-wide computational approach to identify disease-associated mutations derived from IGC events. Our approach revealed hundreds of known pathological mutations that could have been caused by IGC. Further, we identified several dozen high-confidence cases of inherited disease mutations resulting from IGC in ∼1% of all genes analyzed. About half of the donor sequences associated with such mutations are functional paralogous genes, suggesting that epistatic interactions or differential expression patterns will determine the impact upon fitness of specific substitutions between duplicated genes. In addition, we identified thousands of hitherto undescribed and potentially deleterious mutations that could arise via IGC. Our findings reveal the extent of the impact of interlocus gene conversion upon the spectrum of human inherited disease.
Figures

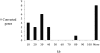

Similar articles
-
Interlocus gene conversion explains at least 2.7% of single nucleotide variants in human segmental duplications.BMC Genomics. 2015 Jun 16;16(1):456. doi: 10.1186/s12864-015-1681-3. BMC Genomics. 2015. PMID: 26077037 Free PMC article.
-
A Phylogenetic Approach Finds Abundant Interlocus Gene Conversion in Yeast.Mol Biol Evol. 2016 Sep;33(9):2469-76. doi: 10.1093/molbev/msw114. Epub 2016 Jun 13. Mol Biol Evol. 2016. PMID: 27297467 Free PMC article.
-
Interlocus Gene Conversion, Natural Selection, and Paralog Homogenization.Mol Biol Evol. 2023 Sep 1;40(9):msad198. doi: 10.1093/molbev/msad198. Mol Biol Evol. 2023. PMID: 37675606 Free PMC article.
-
The mutational spectrum of single base-pair substitutions causing human genetic disease: patterns and predictions.Hum Genet. 1990 Jun;85(1):55-74. doi: 10.1007/BF00276326. Hum Genet. 1990. PMID: 2192981 Review.
-
Charcot-Marie-Tooth disease and related inherited neuropathies.Medicine (Baltimore). 1996 Sep;75(5):233-50. doi: 10.1097/00005792-199609000-00001. Medicine (Baltimore). 1996. PMID: 8862346 Review.
Cited by
-
Biased gene conversion skews allele frequencies in human populations, increasing the disease burden of recessive alleles.Am J Hum Genet. 2014 Oct 2;95(4):408-20. doi: 10.1016/j.ajhg.2014.09.008. Am J Hum Genet. 2014. PMID: 25279983 Free PMC article.
-
A 25-year odyssey of genomic technology advances and structural variant discovery.Cell. 2024 Feb 29;187(5):1024-1037. doi: 10.1016/j.cell.2024.01.002. Epub 2024 Jan 29. Cell. 2024. PMID: 38290514 Free PMC article. Review.
-
Gene Conversion Facilitates the Adaptive Evolution of Self-Resistance in Highly Toxic Newts.Mol Biol Evol. 2021 Sep 27;38(10):4077-4094. doi: 10.1093/molbev/msab182. Mol Biol Evol. 2021. PMID: 34129031 Free PMC article.
-
Robust and accurate estimation of paralog-specific copy number for duplicated genes using whole-genome sequencing.Nat Commun. 2022 Jun 9;13(1):3221. doi: 10.1038/s41467-022-30930-3. Nat Commun. 2022. PMID: 35680869 Free PMC article.
-
Interplay of interlocus gene conversion and crossover in segmental duplications under a neutral scenario.G3 (Bethesda). 2014 Jun 6;4(8):1479-89. doi: 10.1534/g3.114.012435. G3 (Bethesda). 2014. PMID: 24906640 Free PMC article.
References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ 1990. Basic local alignment search tool. J Mol Biol 215: 403–410 - PubMed
-
- Ball EV, Stenson PD, Abeysinghe SS, Krawczak M, Cooper DN, Chuzhanova NA 2005. Microdeletions and microinsertions causing human genetic disease: common mechanisms of mutagenesis and the role of local DNA sequence complexity. Hum Mutat 26: 205–213 - PubMed
-
- Benovoy D, Drouin G 2009. Ectopic gene conversions in the human genome. Genomics 93: 27–32 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
