Accuracy of various human NAT2 SNP genotyping panels to infer rapid, intermediate and slow acetylator phenotypes
- PMID: 22092036
- PMCID: PMC3285565
- DOI: 10.2217/pgs.11.122
Accuracy of various human NAT2 SNP genotyping panels to infer rapid, intermediate and slow acetylator phenotypes
Abstract
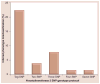
Aim: Humans exhibit genetic polymorphism in NAT2 resulting in rapid, intermediate and slow acetylator phenotypes. Over 65 NAT2 variants possessing one or more SNPs in the 870-bp NAT2 coding region have been reported. The seven most frequent SNPs are rs1801279 (191G>A), rs1041983 (282C>T), rs1801280 (341T>C), rs1799929 (481C>T), rs1799930 (590G>A), rs1208 (803A>G) and rs1799931 (857G>A). The majority of studies investigate the NAT2 genotype assay for three SNPs: 481C>T, 590G>A and 857G>A. A tag-SNP (rs1495741) recently identified in a genome-wide association study has also been proposed as a biomarker for the NAT2 phenotype.
Materials & methods: Sulfamethazine N-acetyltransferase catalytic activities were measured in cryopreserved human hepatocytes from a convenience sample of individuals in the USA with an ethnic frequency similar to the 2010 US population census. These activities were segregated by the tag-SNP rs1495741 and each of the seven SNPs described above. We assessed the accuracy of the tag-SNP and various two-, three-, four- and seven-SNP genotyping panels for their ability to accurately infer NAT2 phenotype.
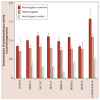
Results: The accuracy of the various NAT2 SNP genotype panels to infer NAT2 phenotype were as follows: seven-SNP: 98.4%; tag-SNP: 77.7%; two-SNP: 96.1%; three-SNP: 92.2%; and four-SNP: 98.4%.
Conclusion: A NAT2 four-SNP genotype panel of rs1801279 (191G>A), rs1801280 (341T>C), rs1799930 (590G>A) and rs1799931 (857G>A) infers NAT2 acetylator phenotype with high accuracy, and is recommended over the tag-, two-, three- and (for economy of scale) the seven-SNP genotyping panels, particularly in populations of non-European ancestry.
Figures



Comment in
-
Accuracy of NAT2 SNP genotyping panels to infer acetylator phenotypes in African, Asian, Amerindian and admixed populations.Pharmacogenomics. 2012 Jun;13(8):851-4; author reply 855. doi: 10.2217/pgs.12.48. Pharmacogenomics. 2012. PMID: 22676187 No abstract available.
References
-
- Weber WW, Hein DW. N-acetylation pharmacogenetics. Pharmacol Rev. 1985;37:25–79. - PubMed
-
- Cascorbi I, Brockmoller J, Mrozikiewicz PM, Muller A, Roots I. Arylamine N-acetyltransferase activity in man. Drug Metab Rev. 1999;31:489–502. - PubMed
-
- Blum M, Demierre A, Grant DM, Heim M, Meyer UA. Molecular mechanism of slow acetylation of drugs and carcinogens in humans. Proc Natl Acad Sci USA. 1991;88:5237–5241. Described the initial report into the functional effects of three common NAT2 SNPs and formed the basis for the historical three-SNP assay. - PMC - PubMed
Websites
-
- Department of pharmacology and toxicology. University of Louisville; KY, USA: www.n-acetyltransferasenomenclature.louisville.edu Official website for consensus human arylamine N-acetyltransferase gene nomenclature.
-
- Celsis Characterization tables. www.celsis.com/ivt/characterization-tables/
-
- US Census Burea. Overview of race and Hispanic origin. 2010 www.census.gov/prod/cen2010/briefs/c2010br-02.pdf.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
