Changes of lipid domains in Bacillus subtilis cells with disrupted cell wall peptidoglycan
- PMID: 22092867
- PMCID: PMC3433793
- DOI: 10.1111/j.1574-6968.2011.02417.x
Changes of lipid domains in Bacillus subtilis cells with disrupted cell wall peptidoglycan
Abstract
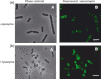
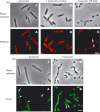
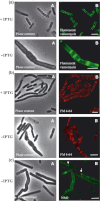
The cell wall is responsible for cell integrity and the maintenance of cell shape in bacteria. The Gram-positive bacterial cell wall consists of a thick peptidoglycan layer located on the outside of the cytoplasmic membrane. Bacterial cell membranes, like eukaryotic cell membranes, are known to contain domains of specific lipid and protein composition. Recently, using the membrane-binding fluorescent dye FM4-64, helix-like lipid structures extending along the long axis of the cell and consisting of negatively charged phospholipids were detected in the rod-shaped bacterium Bacillus subtilis. It was also shown that the cardiolipin-specific dye, nonyl acridine orange (NAO), is preferentially distributed at the cell poles and in the septal regions in both Escherichia coli and B. subtilis. These results suggest that phosphatidylglycerol is the principal component of the observed spiral domains in B. subtilis. Here, using the fluorescent dyes FM4-64 and NAO, we examined whether these lipid domains are linked to the presence of cell wall peptidoglycan. We show that in protoplasted cells, devoid of the peptidoglycan layer, helix-like lipid structures are not preserved. Specific lipid domains are also missing in cells depleted of MurG, an enzyme involved in peptidoglycan synthesis, indicating a link between lipid domain formation and peptidoglycan synthesis.
© 2011 Federation of European Microbiological Societies. Published by Blackwell Publishing Ltd. All rights reserved.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

