Variance decomposition of protein profiles from antibody arrays using a longitudinal twin model
- PMID: 22093360
- PMCID: PMC3247853
- DOI: 10.1186/1477-5956-9-73
Variance decomposition of protein profiles from antibody arrays using a longitudinal twin model
Abstract
Background: The advent of affinity-based proteomics technologies for global protein profiling provides the prospect of finding new molecular biomarkers for common, multifactorial disorders. The molecular phenotypes obtained from studies on such platforms are driven by multiple sources, including genetic, environmental, and experimental components. In characterizing the contribution of different sources of variation to the measured phenotypes, the aim is to facilitate the design and interpretation of future biomedical studies employing exploratory and multiplexed technologies. Thus, biometrical genetic modelling of twin or other family data can be used to decompose the variation underlying a phenotype into biological and experimental components.
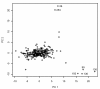
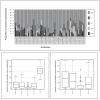
Results: Using antibody suspension bead arrays and antibodies from the Human Protein Atlas, we study unfractionated serum from a longitudinal study on 154 twins. In this study, we provide a detailed description of how the variation in a molecular phenotype in terms of protein profile can be decomposed into familial i.e. genetic and common environmental; individual environmental, short-term biological and experimental components. The results show that across 69 antibodies analyzed in the study, the median proportion of the total variation explained by familial sources is 12% (IQR 1-22%), and the median proportion of the total variation attributable to experimental sources is 63% (IQR 53-72%).
Conclusion: The variability analysis of antibody arrays highlights the importance to consider variability components and their relative contributions when designing and evaluating studies for biomarker discoveries with exploratory, high-throughput and multiplexed methods.
Figures




References
-
- Atkinson AJ, Colburn WA, DeGruttola VG, DeMets DL, Downing GJ, Hoth DF, Oates JA, Peck CC, Schooley RT, Spilker BA. et al.Biomarkers and surrogate endpoints: Preferred definitions and conceptual framework. Clinical Pharmacology & Therapeutics. 2001;69:89–95. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

