Revisiting the folding kinetics of bacteriorhodopsin
- PMID: 22095725
- PMCID: PMC3323784
- DOI: 10.1002/pro.766
Revisiting the folding kinetics of bacteriorhodopsin
Abstract
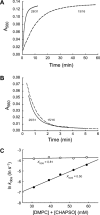
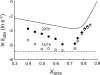
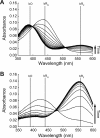
The elucidation of the physical principles that govern the folding and stability of membrane proteins is one of the greatest challenges in protein science. Several insights into the folding of α-helical membrane proteins have come from the investigation of the conformational equilibrium of H. halobium bacteriorhodopsin (bR) in mixed micelles using SDS as a denaturant. In an effort to confirm that folded bR and SDS-denatured bR reach the same conformational equilibrium, we found that bR folding is significantly slower than has been previously known. Interrogation of the effect of the experimental variables on folding kinetics reveals that the rate of folding is dependent not only on the mole fraction of SDS but also on the molar concentrations of mixed micelle components, a variable that was not controlled in the previous study of bR folding kinetics. Moreover, when the molar concentrations of mixed micelle components are fixed at the concentrations commonly employed for bR equilibrium studies, conformational relaxation in the transition zone is slower than hydrolysis of the retinal Schiff base. As a result, the conformational equilibrium between folded bR and SDS-denatured bR cannot be achieved under the conventional condition. Our finding suggests that the molar concentrations of mixed micelle components are important experimental variables in the investigation of the kinetics and thermodynamics of bR folding and should be accounted for to ensure the accurate assessment of the conformational equilibrium of bR without the interference of retinal hydrolysis.
Copyright © 2011 The Protein Society.
Figures


Similar articles
-
Structure and function in bacteriorhodopsin: the role of the interhelical loops in the folding and stability of bacteriorhodopsin.J Mol Biol. 2001 Apr 27;308(2):409-22. doi: 10.1006/jmbi.2001.4603. J Mol Biol. 2001. PMID: 11327776
-
Structure and function in bacteriorhodopsin: the effect of the interhelical loops on the protein folding kinetics.J Mol Biol. 2001 Apr 27;308(2):423-35. doi: 10.1006/jmbi.2001.4604. J Mol Biol. 2001. PMID: 11327777
-
Bicelle size modulates the rate of bacteriorhodopsin folding.Protein Sci. 2018 Jun;27(6):1109-1112. doi: 10.1002/pro.3414. Epub 2018 Apr 25. Protein Sci. 2018. PMID: 29604129 Free PMC article.
-
Probing membrane protein unfolding with pulse proteolysis.J Mol Biol. 2011 Mar 4;406(4):545-51. doi: 10.1016/j.jmb.2010.12.018. Epub 2010 Dec 28. J Mol Biol. 2011. PMID: 21192947 Free PMC article. Review.
-
Retinal proteins as model systems for membrane protein folding.Biochim Biophys Acta. 2014 May;1837(5):656-63. doi: 10.1016/j.bbabio.2013.11.021. Epub 2013 Dec 12. Biochim Biophys Acta. 2014. PMID: 24333783 Review.
Cited by
-
Folding and Misfolding of Human Membrane Proteins in Health and Disease: From Single Molecules to Cellular Proteostasis.Chem Rev. 2019 May 8;119(9):5537-5606. doi: 10.1021/acs.chemrev.8b00532. Epub 2019 Jan 4. Chem Rev. 2019. PMID: 30608666 Free PMC article.
-
Thermostabilization of the β1-adrenergic receptor correlates with increased entropy of the inactive state.J Phys Chem B. 2013 Jun 20;117(24):7283-91. doi: 10.1021/jp403207c. Epub 2013 Jun 5. J Phys Chem B. 2013. PMID: 23697892 Free PMC article.
-
Measuring membrane protein stability under native conditions.Proc Natl Acad Sci U S A. 2014 Jan 7;111(1):219-24. doi: 10.1073/pnas.1318576111. Epub 2013 Dec 23. Proc Natl Acad Sci U S A. 2014. PMID: 24367094 Free PMC article.
-
Cooperative folding of a polytopic α-helical membrane protein involves a compact N-terminal nucleus and nonnative loops.Proc Natl Acad Sci U S A. 2015 Jun 30;112(26):7978-83. doi: 10.1073/pnas.1424751112. Epub 2015 Jun 8. Proc Natl Acad Sci U S A. 2015. PMID: 26056273 Free PMC article.
-
Reversible folding of human peripheral myelin protein 22, a tetraspan membrane protein.Biochemistry. 2013 May 14;52(19):3229-41. doi: 10.1021/bi301635f. Epub 2013 May 2. Biochemistry. 2013. PMID: 23639031 Free PMC article.
References
-
- Fagerberg L, Jonasson K, von Heijne G, Uhlen M, Berglund L. Prediction of the human membrane proteome. Proteomics. 2010;10:1141–1149. - PubMed
-
- Yildirim MA, Goh KI, Cusick ME, Barabasi AL, Vidal M. Drug-target network. Nat Biotechnol. 2007;25:1119–1126. - PubMed
-
- Bowie JU. Solving the membrane protein folding problem. Nature. 2005;438:581–589. - PubMed
-
- Stanley AM, Fleming KG. The process of folding proteins into membranes: challenges and progress. Arch Biochem Biophys. 2008;469:46–66. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

