Modeling large regions in proteins: applications to loops, termini, and folding
- PMID: 22095743
- PMCID: PMC3323786
- DOI: 10.1002/pro.767
Modeling large regions in proteins: applications to loops, termini, and folding
Abstract
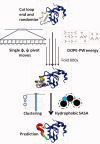
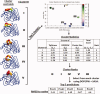
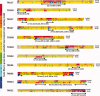
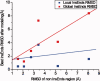
Template-based methods for predicting protein structure provide models for a significant portion of the protein but often contain insertions or chain ends (InsEnds) of indeterminate conformation. The local structure prediction "problem" entails modeling the InsEnds onto the rest of the protein. A well-known limit involves predicting loops of ≤12 residues in crystal structures. However, InsEnds may contain as many as ~50 amino acids, and the template-based model of the protein itself may be imperfect. To address these challenges, we present a free modeling method for predicting the local structure of loops and large InsEnds in both crystal structures and template-based models. The approach uses single amino acid torsional angle "pivot" moves of the protein backbone with a C(β) level representation. Nevertheless, our accuracy for loops is comparable to existing methods. We also apply a more stringent test, the blind structure prediction and refinement categories of the CASP9 tournament, where we improve the quality of several homology based models by modeling InsEnds as long as 45 amino acids, sizes generally inaccessible to existing loop prediction methods. Our approach ranks as one of the best in the CASP9 refinement category that involves improving template-based models so that they can function as molecular replacement models to solve the phase problem for crystallographic structure determination.
Copyright © 2011 The Protein Society.
Figures



References
-
- Moult J. A decade of CASP: progress, bottlenecks and prognosis in protein structure prediction. Curr Opin Struct Biol. 2005;15:285–289. - PubMed
-
- Jones S, Thornton JM. Prediction of protein-protein interaction sites using patch analysis. J Mol Biol. 1997;272:133–143. - PubMed
-
- Wu SJ, Dean DH. Functional significance of loops in the receptor binding domain of Bacillus thuringiensis CryIIIA delta-endotoxin. J Mol Biol. 1996;255:628–640. - PubMed
-
- Bruccoleri RE, Karplus M. Chain closure with bond angle variations. Macromolecules. 1985;18:2767–2773.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

