Role of SPI-1 secreted effectors in acute bovine response to Salmonella enterica Serovar Typhimurium: a systems biology analysis approach
- PMID: 22096503
- PMCID: PMC3214023
- DOI: 10.1371/journal.pone.0026869
Role of SPI-1 secreted effectors in acute bovine response to Salmonella enterica Serovar Typhimurium: a systems biology analysis approach
Abstract
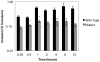
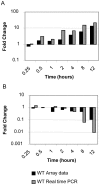
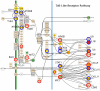
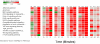
Salmonella enterica Serovar Typhimurium (S. Typhimurium) causes enterocolitis with diarrhea and polymorphonuclear cell (PMN) influx into the intestinal mucosa in humans and calves. The Salmonella Type III Secretion System (T3SS) encoded at Pathogenicity Island I translocates Salmonella effector proteins SipA, SopA, SopB, SopD, and SopE2 into epithelial cells and is required for induction of diarrhea. These effector proteins act together to induce intestinal fluid secretion and transcription of C-X-C chemokines, recruiting PMNs to the infection site. While individual molecular interactions of the effectors with cultured host cells have been characterized, their combined role in intestinal fluid secretion and inflammation is less understood. We hypothesized that comparison of the bovine intestinal mucosal response to wild type Salmonella and a SipA, SopABDE2 effector mutant relative to uninfected bovine ileum would reveal heretofore unidentified diarrhea-associated host cellular pathways. To determine the coordinated effects of these virulence factors, a bovine ligated ileal loop model was used to measure responses to wild type S. Typhimurium (WT) and a ΔsipA, sopABDE2 mutant (MUT) across 12 hours of infection using a bovine microarray. Data were analyzed using standard microarray analysis and a dynamic bayesian network modeling approach (DBN). Both analytical methods confirmed increased expression of immune response genes to Salmonella infection and novel gene expression. Gene expression changes mapped to 219 molecular interaction pathways and 1620 gene ontology groups. Bayesian network modeling identified effects of infection on several interrelated signaling pathways including MAPK, Phosphatidylinositol, mTOR, Calcium, Toll-like Receptor, CCR3, Wnt, TGF-β, and Regulation of Actin Cytoskeleton and Apoptosis that were used to model of host-pathogen interactions. Comparison of WT and MUT demonstrated significantly different patterns of host response at early time points of infection (15 minutes, 30 minutes and one hour) within phosphatidylinositol, CCR3, Wnt, and TGF-β signaling pathways and the regulation of actin cytoskeleton pathway.
Conflict of interest statement
Figures




Similar articles
-
The Salmonella enterica serotype typhimurium effector proteins SipA, SopA, SopB, SopD, and SopE2 act in concert to induce diarrhea in calves.Infect Immun. 2002 Jul;70(7):3843-55. doi: 10.1128/IAI.70.7.3843-3855.2002. Infect Immun. 2002. PMID: 12065528 Free PMC article.
-
SipA, SopA, SopB, SopD, and SopE2 contribute to Salmonella enterica serotype typhimurium invasion of epithelial cells.Infect Immun. 2005 Jan;73(1):146-54. doi: 10.1128/IAI.73.1.146-154.2005. Infect Immun. 2005. PMID: 15618149 Free PMC article.
-
The attenuated sopB mutant of Salmonella enterica serovar Typhimurium has the same tissue distribution and host chemokine response as the wild type in bovine Peyer's patches.Vet Microbiol. 2003 Dec 30;97(3-4):269-77. doi: 10.1016/j.vetmic.2003.09.019. Vet Microbiol. 2003. PMID: 14654296
-
Salmonella effectors: important players modulating host cell function during infection.Cell Microbiol. 2011 Dec;13(12):1858-69. doi: 10.1111/j.1462-5822.2011.01701.x. Epub 2011 Oct 10. Cell Microbiol. 2011. PMID: 21902796 Free PMC article. Review.
-
Salmonella effector proteins and host-cell responses.Cell Mol Life Sci. 2011 Nov;68(22):3687-97. doi: 10.1007/s00018-011-0841-0. Epub 2011 Oct 9. Cell Mol Life Sci. 2011. PMID: 21984608 Free PMC article. Review.
Cited by
-
Systems Biology Analysis of Temporal In vivo Brucella melitensis and Bovine Transcriptomes Predicts host:Pathogen Protein-Protein Interactions.Front Microbiol. 2017 Jul 27;8:1275. doi: 10.3389/fmicb.2017.01275. eCollection 2017. Front Microbiol. 2017. PMID: 28798726 Free PMC article.
-
Outer membrane vesicles in service as protein shuttles, biotic defenders, and immunological doppelgängers.Gut Microbes. 2016 Sep 2;7(5):450-4. doi: 10.1080/19490976.2016.1222345. Epub 2016 Aug 15. Gut Microbes. 2016. PMID: 27600586 Free PMC article. Review.
-
Transcriptome analysis of HeLa cells response to Brucella melitensis infection: a molecular approach to understand the role of the mucosal epithelium in the onset of the Brucella pathogenesis.Microbes Infect. 2012 Aug;14(9):756-67. doi: 10.1016/j.micinf.2012.03.003. Epub 2012 Mar 21. Microbes Infect. 2012. PMID: 22484383 Free PMC article.
-
Reconstruction of the temporal signaling network in Salmonella-infected human cells.Front Microbiol. 2015 Jul 20;6:730. doi: 10.3389/fmicb.2015.00730. eCollection 2015. Front Microbiol. 2015. PMID: 26257716 Free PMC article.
-
Animal models to study acute and chronic intestinal inflammation in mammals.Gut Pathog. 2015 Nov 10;7:29. doi: 10.1186/s13099-015-0076-y. eCollection 2015. Gut Pathog. 2015. PMID: 26561503 Free PMC article. Review.
References
-
- Preliminary FoodNet data on the incidence of infection with pathogens transmitted commonly through food–10 states, 2007. Centers for Disease Control and Prevention MMWR, Morb Mortal Wkly Rep. 2008;57:366–370. - PubMed
-
- Preliminary FoodNet data on the incidence of infection with pathogens transmitted commonly through food - 10 states, 2009. Centers for Disease Control and Prevention MMWR, Morb Mortal Wkly Rep. 2010;59:418–422. - PubMed
-
- Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, et al. The global burden of nontyphoidal Salmonella gastroenteritis. Clin Infect Dis. 2010;50:882–889. - PubMed
-
- Gordon MA, Graham SM. Invasive salmonellosis in Malawi. J Infect Dev Ctries. 2008;2:438–442. - PubMed
-
- Gordon MA, Graham SM, Walsh AL, Wilson L, Phiri A, et al. Epidemics of invasive Salmonella enterica Serovar Enteritidis and S. enterica Serovar Typhimurium infection associated with multidrug resistance among adults and children in Malawi. Clin Infect Dis. 2008;46:963–969. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

