Defining the role of essential genes in human disease
- PMID: 22096564
- PMCID: PMC3214036
- DOI: 10.1371/journal.pone.0027368
Defining the role of essential genes in human disease
Abstract
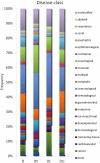
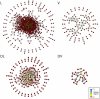
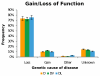
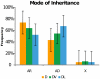
A greater understanding of the causes of human disease can come from identifying characteristics that are specific to disease genes. However, a full understanding of the contribution of essential genes to human disease is lacking, due to the premise that these genes tend to cause developmental abnormalities rather than adult disease. We tested the hypothesis that human orthologs of mouse essential genes are associated with a variety of human diseases, rather than only those related to miscarriage and birth defects. We segregated human disease genes according to whether the knockout phenotype of their mouse ortholog was lethal or viable, defining those with orthologs producing lethal knockouts as essential disease genes. We show that the human orthologs of mouse essential genes are associated with a wide spectrum of diseases affecting diverse physiological systems. Notably, human disease genes with essential mouse orthologs are over-represented among disease genes associated with cancer, suggesting links between adult cellular abnormalities and developmental functions. The proteins encoded by essential genes are highly connected in protein-protein interaction networks, which we find correlates with an over-representation of nuclear proteins amongst essential disease genes. Disease genes associated with essential orthologs also are more likely than those with non-essential orthologs to contribute to disease through an autosomal dominant inheritance pattern, suggesting that these diseases may actually result from semi-dominant mutant alleles. Overall, we have described attributes found in disease genes according to the essentiality status of their mouse orthologs. These findings demonstrate that disease genes do occupy highly connected positions in protein-protein interaction networks, and that due to the complexity of disease-associated alleles, essential genes cannot be ignored as candidates for causing diverse human diseases.
Conflict of interest statement
Figures
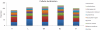
Similar articles
-
Identifying mouse developmental essential genes using machine learning.Dis Model Mech. 2018 Dec 13;11(12):dmm034546. doi: 10.1242/dmm.034546. Dis Model Mech. 2018. PMID: 30563825 Free PMC article.
-
Analysis of human disease genes in the context of gene essentiality.Genomics. 2008 Dec;92(6):414-8. doi: 10.1016/j.ygeno.2008.08.001. Epub 2008 Oct 1. Genomics. 2008. PMID: 18786629
-
Lethal phenotypes in Mendelian disorders.Genet Med. 2024 Jul;26(7):101141. doi: 10.1016/j.gim.2024.101141. Epub 2024 Apr 15. Genet Med. 2024. PMID: 38629401 Free PMC article.
-
Gene discovery informatics toolkit defines candidate genes for unexplained infertility and prenatal or infantile mortality.NPJ Genom Med. 2019 Apr 15;4:8. doi: 10.1038/s41525-019-0081-z. eCollection 2019. NPJ Genom Med. 2019. PMID: 30993004 Free PMC article. Review.
-
Resurrection from lethal knockouts: Bypass of gene essentiality.Biochem Biophys Res Commun. 2020 Jul 30;528(3):405-412. doi: 10.1016/j.bbrc.2020.05.207. Epub 2020 Jun 4. Biochem Biophys Res Commun. 2020. PMID: 32507598 Review.
Cited by
-
Local Action with Global Impact: Highly Similar Infection Patterns of Human Viruses and Bacteriophages.mSystems. 2016 Mar 8;1(2):e00030-15. doi: 10.1128/mSystems.00030-15. eCollection 2016 Mar-Apr. mSystems. 2016. PMID: 27822522 Free PMC article.
-
Essential genes: a cross-species perspective.Mamm Genome. 2023 Sep;34(3):357-363. doi: 10.1007/s00335-023-09984-1. Epub 2023 Mar 10. Mamm Genome. 2023. PMID: 36897351 Free PMC article. Review.
-
Identification of discriminant features from stationary pattern of nucleotide bases and their application to essential gene classification.Front Genet. 2023 Apr 20;14:1154120. doi: 10.3389/fgene.2023.1154120. eCollection 2023. Front Genet. 2023. PMID: 37152988 Free PMC article.
-
Effects of different kinds of essentiality on sequence evolution of human testis proteins.Sci Rep. 2017 Mar 8;7:43534. doi: 10.1038/srep43534. Sci Rep. 2017. PMID: 28272493 Free PMC article.
-
Lost in translocation: the functions of the 18-kD translocator protein.Trends Endocrinol Metab. 2015 Jul;26(7):349-56. doi: 10.1016/j.tem.2015.04.001. Epub 2015 May 28. Trends Endocrinol Metab. 2015. PMID: 26026242 Free PMC article. Review.
References
-
- Smith NG, Eyre-Walker A. Human disease genes: patterns and predictions. Gene. 2003;318:169–175. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

