SCF ubiquitin ligases in the maintenance of genome stability
- PMID: 22099186
- PMCID: PMC3278546
- DOI: 10.1016/j.tibs.2011.10.004
SCF ubiquitin ligases in the maintenance of genome stability
Abstract
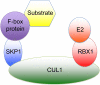
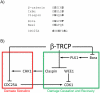
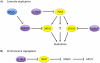
In response to genotoxic stress, eukaryotic cells activate the DNA damage response (DDR), a series of pathways that coordinate cell cycle arrest and DNA repair to prevent deleterious mutations. In addition, cells possess checkpoint mechanisms that prevent aneuploidy by regulating the number of centrosomes and spindle assembly. Among these mechanisms, ubiquitin-mediated degradation of key proteins has an important role in the regulation of the DDR, centrosome duplication and chromosome segregation. This review discusses the functions of a group of ubiquitin ligases, the SCF (SKP1-CUL1-F-box protein) family, in the maintenance of genome stability. Given that general proteasome inhibitors are currently used as anticancer agents, a better understanding of the ubiquitylation of specific targets by specific ubiquitin ligases may result in improved cancer therapeutics.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures
References
-
- Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. - PubMed
-
- Hoare M, et al. Ageing, telomeres, senescence, and liver injury. J Hepatol. 2010;53:950–961. - PubMed
-
- Abraham RT. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 2001;15:2177–2196. - PubMed
-
- Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat Rev Cancer. 2003;3:155–168. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

