RAM/Fam103a1 is required for mRNA cap methylation
- PMID: 22099306
- PMCID: PMC3235549
- DOI: 10.1016/j.molcel.2011.08.041
RAM/Fam103a1 is required for mRNA cap methylation
Abstract
The 7-methylguanosine cap added to the 5' end of mRNA is required for efficient gene expression in eukaryotes. In mammals, methylation of the guanosine cap is catalyzed by RNMT (RNA guanine-7 methyltransferase), an enzyme previously thought to function as a monomer. We have identified an obligate component of the mammalian cap methyltransferase, RAM (RNMT-Activating Mini protein)/Fam103a1, a previously uncharacterized protein. RAM consists of an N-terminal RNMT-activating domain and a C-terminal RNA-binding domain. As monomers RNMT and RAM have a relatively weak affinity for RNA; however, together their RNA affinity is significantly increased. RAM is required for efficient cap methylation in vitro and in vivo, and is indirectly required to maintain mRNA expression levels, for mRNA translation and for cell viability. Our findings demonstrate that RAM is an essential component of the core gene expression machinery.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures


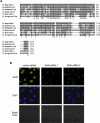
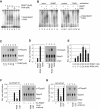
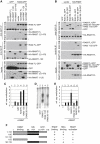
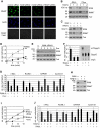


Similar articles
-
Mechanism of allosteric activation of human mRNA cap methyltransferase (RNMT) by RAM: insights from accelerated molecular dynamics simulations.Nucleic Acids Res. 2019 Sep 19;47(16):8675-8692. doi: 10.1093/nar/gkz613. Nucleic Acids Res. 2019. PMID: 31329932 Free PMC article.
-
RAM function is dependent on Kapβ2-mediated nuclear entry.Biochem J. 2014 Feb 1;457(3):473-84. doi: 10.1042/BJ20131359. Biochem J. 2014. PMID: 24200467 Free PMC article.
-
c-Myc co-ordinates mRNA cap methylation and ribosomal RNA production.Biochem J. 2017 Feb 1;474(3):377-384. doi: 10.1042/BCJ20160930. Epub 2016 Dec 1. Biochem J. 2017. PMID: 27934633 Free PMC article.
-
The RNA cap methyltransferases RNMT and CMTR1 co-ordinate gene expression during neural differentiation.Biochem Soc Trans. 2023 Jun 28;51(3):1131-1141. doi: 10.1042/BST20221154. Biochem Soc Trans. 2023. PMID: 37145036 Free PMC article. Review.
-
Regulation of mRNA cap methylation.Biochem J. 2009 Dec 23;425(2):295-302. doi: 10.1042/BJ20091352. Biochem J. 2009. PMID: 20025612 Free PMC article. Review.
Cited by
-
RNA methyltransferases involved in 5' cap biosynthesis.RNA Biol. 2014;11(12):1597-607. doi: 10.1080/15476286.2015.1004955. RNA Biol. 2014. PMID: 25626080 Free PMC article. Review.
-
The End of a 60-year Riddle: Identification and Genomic Characterization of an Iridovirus, the Causative Agent of White Fat Cell Disease in Zooplankton.G3 (Bethesda). 2018 Mar 28;8(4):1259-1272. doi: 10.1534/g3.117.300429. G3 (Bethesda). 2018. PMID: 29487186 Free PMC article.
-
mRNAs of plants and green algae lack the m7G cap-1 structure.New Phytol. 2025 Apr;246(2):396-401. doi: 10.1111/nph.70033. Epub 2025 Feb 27. New Phytol. 2025. PMID: 40016992 Free PMC article. No abstract available.
-
mRNA Cap Methyltransferase, RNMT-RAM, Promotes RNA Pol II-Dependent Transcription.Cell Rep. 2018 May 1;23(5):1530-1542. doi: 10.1016/j.celrep.2018.04.004. Cell Rep. 2018. PMID: 29719263 Free PMC article.
-
Mechanism of allosteric activation of human mRNA cap methyltransferase (RNMT) by RAM: insights from accelerated molecular dynamics simulations.Nucleic Acids Res. 2019 Sep 19;47(16):8675-8692. doi: 10.1093/nar/gkz613. Nucleic Acids Res. 2019. PMID: 31329932 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

