NSD2 links dimethylation of histone H3 at lysine 36 to oncogenic programming
- PMID: 22099308
- PMCID: PMC3222870
- DOI: 10.1016/j.molcel.2011.08.042
NSD2 links dimethylation of histone H3 at lysine 36 to oncogenic programming
Abstract
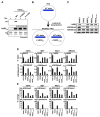
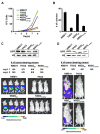
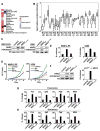
The histone lysine methyltransferase NSD2 (MMSET/WHSC1) is implicated in diverse diseases and commonly overexpressed in multiple myeloma due to a recurrent t(4;14) chromosomal translocation. However, the precise catalytic activity of NSD2 is obscure, preventing progress in understanding how this enzyme influences chromatin biology and myeloma pathogenesis. Here, we show that dimethylation of histone H3 at lysine 36 (H3K36me2) is the principal chromatin-regulatory activity of NSD2. Catalysis of H3K36me2 by NSD2 is sufficient for gene activation. In t(4;14)-positive myeloma cells, the normal genome-wide and gene-specific distribution of H3K36me2 is obliterated, creating a chromatin landscape that selects for a transcription profile favorable for myelomagenesis. Catalytically active NSD2 confers xenograft tumor formation upon t(4;14)-negative cells and promotes oncogenic transformation of primary cells in an H3K36me2-dependent manner. Together, our findings establish H3K36me2 as the primary product generated by NSD2 and demonstrate that genomic disorganization of this canonical chromatin mark by NSD2 initiates oncogenic programming.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures




References
-
- Anderson KC, Carrasco RD. Pathogenesis of myeloma. Annu Rev Pathol. 2011;6:249–274. - PubMed
-
- Bannister AJ, Schneider R, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T. Spatial distribution of di- and tri-methyl lysine 36 of histone H3 at active genes. J Biol Chem. 2005;280:17732–17736. - PubMed
-
- Chesi M, Nardini E, Lim RS, Smith KD, Kuehl WM, Bergsagel PL. The t(4;14) translocation in myeloma dysregulates both FGFR3 and a novel gene, MMSET, resulting in IgH/MMSET hybrid transcripts. Blood. 1998;92:3025–3034. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

