Structural basis for activation of calcineurin by calmodulin
- PMID: 22100452
- PMCID: PMC3258384
- DOI: 10.1016/j.jmb.2011.11.008
Structural basis for activation of calcineurin by calmodulin
Abstract
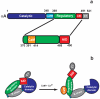
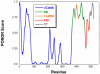
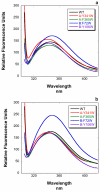
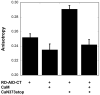
The highly conserved phosphatase calcineurin (CaN) plays vital roles in numerous processes including T-cell activation, development and function of the central nervous system, and cardiac growth. It is activated by the calcium sensor calmodulin (CaM). CaM binds to a regulatory domain (RD) within CaN, causing a conformational change that displaces an autoinhibitory domain (AID) from the active site, resulting in activation of the phosphatase. This is the same general mechanism by which CaM activates CaM-dependent protein kinases. Previously published data have hinted that the RD of CaN is intrinsically disordered. In this work, we demonstrate that the RD is unstructured and that it folds upon binding CaM, ousting the AID from the catalytic site. The RD is 95 residues long, with the AID attached to its C-terminal end and the 24-residue CaM binding region toward the N-terminal end. This is unlike the CaM-dependent protein kinases that have CaM binding sites and AIDs immediately adjacent in sequence. Our data demonstrate that not only does the CaM binding region folds but also an ∼25- to 30-residue region between it and the AID folds, resulting in over half of the RD adopting α-helical structure. This appears to be the first observation of CaM inducing folding of this scale outside of its binding site on a target protein.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures



Similar articles
-
The distal helix in the regulatory domain of calcineurin is important for domain stability and enzyme function.Biochemistry. 2013 Dec 3;52(48):8643-51. doi: 10.1021/bi400483a. Epub 2013 Nov 15. Biochemistry. 2013. PMID: 24191726
-
Structural basis of calcineurin activation by calmodulin.Cell Signal. 2013 Dec;25(12):2661-7. doi: 10.1016/j.cellsig.2013.08.033. Epub 2013 Sep 7. Cell Signal. 2013. PMID: 24018048
-
Calmodulin-Calcineurin Interaction beyond the Calmodulin-Binding Region Contributes to Calcineurin Activation.Biochemistry. 2019 Oct 1;58(39):4070-4085. doi: 10.1021/acs.biochem.9b00626. Epub 2019 Sep 19. Biochemistry. 2019. PMID: 31483613 Free PMC article.
-
Proteins with calmodulin-like domains: structures and functional roles.Cell Mol Life Sci. 2019 Jun;76(12):2299-2328. doi: 10.1007/s00018-019-03062-z. Epub 2019 Mar 15. Cell Mol Life Sci. 2019. PMID: 30877334 Free PMC article. Review.
-
The Merck Frosst Award Lecture 1994. Calmodulin: a versatile calcium mediator protein.Biochem Cell Biol. 1994 Sep-Oct;72(9-10):357-76. Biochem Cell Biol. 1994. PMID: 7605608 Review.
Cited by
-
Stochastic machines as a colocalization mechanism for scaffold protein function.FEBS Lett. 2013 Jun 5;587(11):1587-91. doi: 10.1016/j.febslet.2013.04.006. Epub 2013 Apr 18. FEBS Lett. 2013. PMID: 23603389 Free PMC article.
-
Effects of datumetine on hippocampal NMDAR activity.Toxicol Rep. 2021 May 24;8:1131-1142. doi: 10.1016/j.toxrep.2021.05.009. eCollection 2021. Toxicol Rep. 2021. PMID: 34150523 Free PMC article.
-
Calcineurin Subunits A and B Interact to Regulate Growth and Asexual and Sexual Development in Neurospora crassa.PLoS One. 2016 Mar 28;11(3):e0151867. doi: 10.1371/journal.pone.0151867. eCollection 2016. PLoS One. 2016. PMID: 27019426 Free PMC article.
-
The Recognition of Calmodulin to the Target Sequence of Calcineurin-A Novel Binding Mode.Molecules. 2017 Sep 21;22(10):1584. doi: 10.3390/molecules22101584. Molecules. 2017. PMID: 28934144 Free PMC article.
-
PPP3CA truncating variants clustered in the regulatory domain cause early-onset refractory epilepsy.Clin Genet. 2021 Aug;100(2):227-233. doi: 10.1111/cge.13979. Epub 2021 Jun 1. Clin Genet. 2021. PMID: 33963760 Free PMC article.
References
-
- Wang JH, Desai R. A brain protein and its effect on the Ca2+-and protein modulator-activated cyclic nucleotide phosphodiesterase. Biochem Biophys Res Commun. 1976;72:926–932. - PubMed
-
- Watterson DM, Vanaman TC. Affinity chromatography purification of a cyclic nucleotide phosphodiesterase using immobilized modulator protein, a troponin C-like protein from brain. Biochem Biophys Res Commun. 1976;73:40–46. - PubMed
-
- Klee CB, Krinks MH. Purification of cyclic 3“,5-”nucleotide phosphodiesterase inhibitory protein by affinity chromatography on activator protein coupled to Sepharose. Biochemistry. 1978;17:120–126. - PubMed
-
- Rusnak F, Mertz P. Calcineurin: form and function. Physiol Rev. 2000;80:1483–1521. - PubMed
-
- Chakraborti S, Das S, Kar P, Ghosh B, Samanta K, Kolley S, Ghosh S, Roy S, Chakraborti T. Calcium signaling phenomena in heart diseases: a perspective. Mol Cell Biochem. 2007;298:1–40. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

