Type three effector gene distribution and sequence analysis provide new insights into the pathogenicity of plant-pathogenic Xanthomonas arboricola
- PMID: 22101042
- PMCID: PMC3255760
- DOI: 10.1128/AEM.06119-11
Type three effector gene distribution and sequence analysis provide new insights into the pathogenicity of plant-pathogenic Xanthomonas arboricola
Abstract
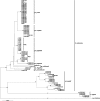
Xanthomonas arboricola is a complex bacterial species which mainly attacks fruit trees and is responsible for emerging diseases in Europe. It comprises seven pathovars (X. arboricola pv. pruni, X. arboricola pv. corylina, X. arboricola pv. juglandis, X. arboricola pv. populi, X. arboricola pv. poinsettiicola, X. arboricola pv. celebensis, and X. arboricola pv. fragariae), each exhibiting characteristic disease symptoms and distinct host specificities. To better understand the factors underlying this ecological trait, we first assessed the phylogenetic relationships among a worldwide collection of X. arboricola strains by sequencing the housekeeping gene rpoD. This analysis revealed that strains of X. arboricola pathovar populi are divergent from the main X. arboricola cluster formed by all other strains. Then, we investigated the distribution of 53 type III effector (T3E) genes in a collection of 57 X. arboricola strains that are representative of the main X. arboricola cluster. Our results showed that T3E repertoires vary greatly between X. arboricola pathovars in terms of size. Indeed, X. arboricola pathovars pruni, corylina, and juglandis, which are responsible for economically important stone fruit and nut diseases in Europe, harbored the largest T3E repertoires, whereas pathovars poinsettiicola, celebensis, and fragariae harbored the smallest. We also identified several differences in T3E gene content between X. arboricola pathovars pruni, corylina, and juglandis which may account for their differing host specificities. Further, we examined the allelic diversity of eight T3E genes from X. arboricola pathovars. This analysis revealed very limited allelic variations at the different loci. Altogether, the data presented here provide new insights into the evolution of pathogenicity and host range of X. arboricola and are discussed in terms of emergence of new diseases within this bacterial species.
Figures


References
-
- Alfano JR, Collmer A. 2004. Type III secretion system effector proteins: double agents in bacterial disease and plant defense. Annu. Rev. Phytopathol. 42:385–414 - PubMed
-
- Al-Saadi A, et al. 2007. All five host-range variants of Xanthomonas citri carry one pthA homolog with 17.5 repeats that determines pathogenicity on citrus, but none determine host-range variation. Mol. Plant Microbe Interact. 20:934–943 - PubMed
-
- . 2000. Council directive 2000/29/EC of 8 May 2000 on protective measures against the introduction into the community of organisms harmful to plants or plant products and against their spread within the Community. Off. J. Eur. Communities L169:1–112
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

