Vanadium nitrogenase: a two-hit wonder?
- PMID: 22101422
- PMCID: PMC3823560
- DOI: 10.1039/c1dt11535a
Vanadium nitrogenase: a two-hit wonder?
Abstract
Nitrogenase catalyzes the biological conversion of atmospheric dinitrogen to bioavailable ammonia. The molybdenum (Mo)- and vanadium (V)-dependent nitrogenases are two homologous members of this metalloenzyme family. However, despite their similarities in structure and function, the characterization of V-nitrogenase has taken a much longer and more winding path than that of its Mo-counterpart. From the initial discovery of this nitrogen-fixing system, to the recent finding of its CO-reducing capacity, V-nitrogenase has proven to be a two-hit wonder in the over-a-century-long research of nitrogen fixation. This perspective provides a brief account of the catalytic function and structural basis of V-nitrogenase, as well as a short discussion of the theoretical and practical potentials of this unique metalloenzyme.
Figures




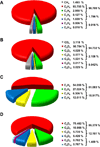

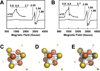
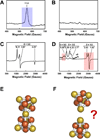
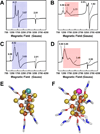

References
-
- Burgess BK, Lowe DJ. Chem. Rev. 1996;96:2983–3012. - PubMed
-
- Georgiadis MM, Komiya H, Chakrabarti P, Woo D, Kornuc JJ, Rees DC. Science. 1992;257:1653–1659. - PubMed
-
- Einsle O, Tezcan FA, Andrade SL, Schmid B, Yoshida M, Howard JB, Rees DC. Science. 2002;297:1696–1700. - PubMed
-
- Schindelin H, Kisker C, Schlessman JL, Howard JB, Rees DC. Nature. 1997;387:370–376. - PubMed
-
- Bishop PE, Premakumar R. In: Biological Nitorgen Fixation. Stacey G, Burris RH, Evans HJ, editors. New York: Chapman and Hall; 1992. pp. 736–762.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

