Crosstalk between B lymphocytes, microbiota and the intestinal epithelium governs immunity versus metabolism in the gut
- PMID: 22101768
- PMCID: PMC3902046
- DOI: 10.1038/nm.2505
Crosstalk between B lymphocytes, microbiota and the intestinal epithelium governs immunity versus metabolism in the gut
Abstract
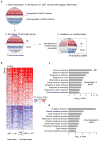
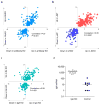
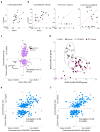
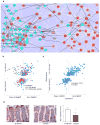
Using a systems biology approach, we discovered and dissected a three-way interaction between the immune system, the intestinal epithelium and the microbiota. We found that, in the absence of B cells, or of IgA, and in the presence of the microbiota, the intestinal epithelium launches its own protective mechanisms, upregulating interferon-inducible immune response pathways and simultaneously repressing Gata4-related metabolic functions. This shift in intestinal function leads to lipid malabsorption and decreased deposition of body fat. Network analysis revealed the presence of two interconnected epithelial-cell gene networks, one governing lipid metabolism and another regulating immunity, that were inversely expressed. Gene expression patterns in gut biopsies from individuals with common variable immunodeficiency or with HIV infection and intestinal malabsorption were very similar to those of the B cell-deficient mice, providing a possible explanation for a longstanding enigmatic association between immunodeficiency and defective lipid absorption in humans.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

Comment in
-
A gut triumvirate rules homeostasis.Nat Med. 2011 Dec 6;17(12):1549-50. doi: 10.1038/nm.2592. Nat Med. 2011. PMID: 22146454 Free PMC article.
-
Mucosal immunology: Hungry for the fight.Nat Rev Immunol. 2011 Dec 23;12(1):2. doi: 10.1038/nri3136. Nat Rev Immunol. 2011. PMID: 22193762 No abstract available.
References
-
- Cerf-Bensussan N, Gaboriau-Routhiau V. The immune system and the gut microbiota: friends or foes? Nat Rev Immunol. 2010;10:735–744. - PubMed
-
- Hooper LV, Macpherson AJ. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol. 2010;10:159–169. - PubMed
-
- Gaboriau-Routhiau V, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–689. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

