The E3 ubiquitin ligase Rnf8 stabilizes Tpp1 to promote telomere end protection
- PMID: 22101936
- PMCID: PMC3657743
- DOI: 10.1038/nsmb.2172
The E3 ubiquitin ligase Rnf8 stabilizes Tpp1 to promote telomere end protection
Abstract
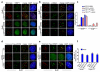
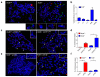
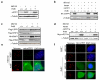
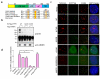
The mammalian shelterin component TPP1 has essential roles in telomere maintenance and, together with POT1, is required for the repression of DNA damage signaling at telomeres. Here we show that in Mus musculus, the E3 ubiquitin ligase Rnf8 localizes to uncapped telomeres and promotes the accumulation of DNA damage proteins 53Bp1 and γ-H2ax. In the absence of Rnf8, Tpp1 is unstable, resulting in telomere shortening and chromosome fusions through the alternative nonhomologous end-joining (A-NHEJ) repair pathway. The Rnf8 RING-finger domain is essential for Tpp1 stability and retention at telomeres. Rnf8 physically interacts with Tpp1 to generate Ubc13-dependent Lys63 polyubiquitin chains that stabilize Tpp1 at telomeres. The conserved Tpp1 residue Lys233 is important for Rnf8-mediated Tpp1 ubiquitylation and localization to telomeres. Thus, Tpp1 is a newly identified substrate for Rnf8, indicating a previously unrecognized role for Rnf8 in telomere end protection.
Figures


References
-
- de Lange T. How Shelterin Solves the Telomere End-Protection Problem. Cold Spring Harb Symp Quant Biol. 2011 - PubMed
-
- Wang F, et al. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature. 2007;445:506–10. - PubMed
-
- Xin H, et al. TPP1 is a homologue of ciliate TEBP-beta and interacts with POT1 to recruit telomerase. Nature. 2007;445:559–62. - PubMed
-
- Denchi EL, de Lange T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature. 2007;448:1068–71. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

