A single autoimmune T cell receptor recognizes more than a million different peptides
- PMID: 22102287
- PMCID: PMC3256900
- DOI: 10.1074/jbc.M111.289488
A single autoimmune T cell receptor recognizes more than a million different peptides
Abstract
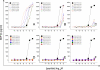
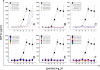
The T cell receptor (TCR) orchestrates immune responses by binding to foreign peptides presented at the cell surface in the context of major histocompatibility complex (MHC) molecules. Effective immunity requires that all possible foreign peptide-MHC molecules are recognized or risks leaving holes in immune coverage that pathogens could quickly evolve to exploit. It is unclear how a limited pool of <10(8) human TCRs can successfully provide immunity to the vast array of possible different peptides that could be produced from 20 proteogenic amino acids and presented by self-MHC molecules (>10(15) distinct peptide-MHCs). One possibility is that T cell immunity incorporates an extremely high level of receptor degeneracy, enabling each TCR to recognize multiple peptides. However, the extent of such TCR degeneracy has never been fully quantified. Here, we perform a comprehensive experimental and mathematical analysis to reveal that a single patient-derived autoimmune CD8(+) T cell clone of pathogenic relevance in human type I diabetes recognizes >one million distinct decamer peptides in the context of a single MHC class I molecule. A large number of peptides that acted as substantially better agonists than the wild-type "index" preproinsulin-derived peptide (ALWGPDPAAA) were identified. The RQFGPDFPTI peptide (sampled from >10(8) peptides) was >100-fold more potent than the index peptide despite differing from this sequence at 7 of 10 positions. Quantification of this previously unappreciated high level of CD8(+) T cell cross-reactivity represents an important step toward understanding the system requirements for adaptive immunity and highlights the enormous potential of TCR degeneracy to be the causative factor in autoimmune disease.
Figures






References
-
- Rudolph M. G., Wilson I. A. (2002) The specificity of TCR/pMHC interaction. Curr. Opin. Immunol. 14, 52–65 - PubMed
-
- Rudolph M. G., Stanfield R. L., Wilson I. A. (2006) How TCRs bind MHCs, peptides, and coreceptors. Annu. Rev. Immunol. 24, 419–466 - PubMed
-
- Mason D. (1998) A very high level of crossreactivity is an essential feature of the T-cell receptor. Immunol. Today 19, 395–404 - PubMed
-
- Arstila T. P., Casrouge A., Baron V., Even J., Kanellopoulos J., Kourilsky P. (1999) A direct estimate of the human αβ T cell receptor diversity. Science 286, 958–961 - PubMed
-
- Wraith D. C., Bruun B., Fairchild P. J. (1992) Cross-reactive antigen recognition by an encephalitogenic T cell receptor. Implications for T cell biology and autoimmunity. J. Immunol. 149, 3765–3770 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

