Cross-reactive T cells are involved in rapid clearance of 2009 pandemic H1N1 influenza virus in nonhuman primates
- PMID: 22102819
- PMCID: PMC3213121
- DOI: 10.1371/journal.ppat.1002381
Cross-reactive T cells are involved in rapid clearance of 2009 pandemic H1N1 influenza virus in nonhuman primates
Abstract
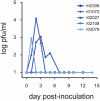
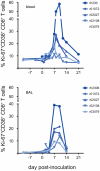
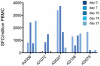
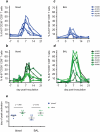
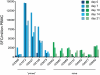
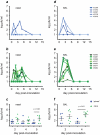
In mouse models of influenza, T cells can confer broad protection against multiple viral subtypes when antibodies raised against a single subtype fail to do so. However, the role of T cells in protecting humans against influenza remains unclear. Here we employ a translational nonhuman primate model to show that cross-reactive T cell responses play an important role in early clearance of infection with 2009 pandemic H1N1 influenza virus (H1N1pdm). To "prime" cellular immunity, we first infected 5 rhesus macaques with a seasonal human H1N1 isolate. These animals made detectable cellular and antibody responses against the seasonal H1N1 isolate but had no neutralizing antibodies against H1N1pdm. Four months later, we challenged the 5 "primed" animals and 7 naive controls with H1N1pdm. In naive animals, CD8+ T cells with an activated phenotype (Ki-67+ CD38+) appeared in blood and lung 5-7 days post inoculation (p.i.) with H1N1pdm and reached peak magnitude 7-10 days p.i. In contrast, activated T cells were recruited to the lung as early as 2 days p.i. in "primed" animals, and reached peak frequencies in blood and lung 4-7 days p.i. Interferon (IFN)-γ Elispot and intracellular cytokine staining assays showed that the virus-specific response peaked earlier and reached a higher magnitude in "primed" animals than in naive animals. This response involved both CD4+ and CD8+ T cells. Strikingly, "primed" animals cleared H1N1pdm infection significantly earlier from the upper and lower respiratory tract than the naive animals did, and before the appearance of H1N1pdm-specific neutralizing antibodies. Together, our results suggest that cross-reactive T cell responses can mediate early clearance of an antigenically novel influenza virus in primates. Vaccines capable of inducing such cross-reactive T cells may help protect humans against severe disease caused by newly emerging pandemic influenza viruses.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- US Centers for Disease Control and Prevention. Serum cross-reactive antibody response to a novel influenza A (H1N1) virus after vaccination with seasonal influenza vaccine. MMWR Morb Mortal Wkly Rep. 2009;58:521–524. - PubMed
-
- Hancock K, Veguilla V, Lu X, Zhong W, Butler EN, et al. Cross-reactive antibody responses to the 2009 pandemic H1N1 influenza virus. N Engl J Med. 2009;361:1945–1952. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

