Relative burden of large CNVs on a range of neurodevelopmental phenotypes
- PMID: 22102821
- PMCID: PMC3213131
- DOI: 10.1371/journal.pgen.1002334
Relative burden of large CNVs on a range of neurodevelopmental phenotypes
Abstract
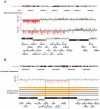
While numerous studies have implicated copy number variants (CNVs) in a range of neurological phenotypes, the impact relative to disease severity has been difficult to ascertain due to small sample sizes, lack of phenotypic details, and heterogeneity in platforms used for discovery. Using a customized microarray enriched for genomic hotspots, we assayed for large CNVs among 1,227 individuals with various neurological deficits including dyslexia (376), sporadic autism (350), and intellectual disability (ID) (501), as well as 337 controls. We show that the frequency of large CNVs (>1 Mbp) is significantly greater for ID-associated phenotypes compared to autism (p = 9.58 × 10(-11), odds ratio = 4.59), dyslexia (p = 3.81 × 10(-18), odds ratio = 14.45), or controls (p = 2.75 × 10(-17), odds ratio = 13.71). There is a striking difference in the frequency of rare CNVs (>50 kbp) in autism (10%, p = 2.4 × 10(-6), odds ratio = 6) or ID (16%, p = 3.55 × 10(-12), odds ratio = 10) compared to dyslexia (2%) with essentially no difference in large CNV burden among dyslexia patients compared to controls. Rare CNVs were more likely to arise de novo (64%) in ID when compared to autism (40%) or dyslexia (0%). We observed a significantly increased large CNV burden in individuals with ID and multiple congenital anomalies (MCA) compared to ID alone (p = 0.001, odds ratio = 2.54). Our data suggest that large CNV burden positively correlates with the severity of childhood disability: ID with MCA being most severely affected and dyslexics being indistinguishable from controls. When autism without ID was considered separately, the increase in CNV burden was modest compared to controls (p = 0.07, odds ratio = 2.33).
Conflict of interest statement
EEE is on the scientific advisory board for Pacific Biosciences.
Figures





References
-
- Sharp AJ, Hansen S, Selzer RR, Cheng Z, Regan R, et al. Discovery of previously unidentified genomic disorders from the duplication architecture of the human genome. Nat Genet. 2006;38:1038–1042. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

