A new piece of the Shigella Pathogenicity puzzle: spermidine accumulation by silencing of the speG gene [corrected]
- PMID: 22102881
- PMCID: PMC3213128
- DOI: 10.1371/journal.pone.0027226
A new piece of the Shigella Pathogenicity puzzle: spermidine accumulation by silencing of the speG gene [corrected]
Erratum in
- PLoS One. 2012;7(5): doi/10.1371/annotation/272940da-34b6-4cd7-8794-ecb5df8d7cc2
Abstract
The genome of Shigella, a gram negative bacterium which is the causative agent of bacillary dysentery, shares strong homologies with that of its commensal ancestor, Escherichia coli. The acquisition, by lateral gene transfer, of a large plasmid carrying virulence determinants has been a crucial event in the evolution towards the pathogenic lifestyle and has been paralleled by the occurrence of mutations affecting genes, which negatively interfere with the expression of virulence factors. In this context, we have analysed to what extent the presence of the plasmid-encoded virF gene, the major activator of the Shigella regulon for invasive phenotype, has modified the transcriptional profile of E. coli. Combining results from transcriptome assays and comparative genome analyses we show that in E. coli VirF, besides being able to up-regulate several chromosomal genes, which potentially influence bacterial fitness within the host, also activates genes which have been lost by Shigella. We have focused our attention on the speG gene, which encodes spermidine acetyltransferase, an enzyme catalysing the conversion of spermidine into the physiologically inert acetylspermidine, since recent evidence stresses the involvement of polyamines in microbial pathogenesis. Through identification of diverse mutations, which prevent expression of a functional SpeG protein, we show that the speG gene has been silenced by convergent evolution and that its inactivation causes the marked increase of intracellular spermidine in all Shigella spp. This enhances the survival of Shigella under oxidative stress and allows it to better face the adverse conditions it encounters inside macrophage. This is supported by the outcome of infection assays performed in mouse peritoneal macrophages and of a competitive-infection assay on J774 macrophage cell culture. Our observations fully support the pathoadaptive nature of speG inactivation in Shigella and reveal that the accumulation of spermidine is a key determinant in the pathogenicity strategy adopted by this microrganism.
Conflict of interest statement
Figures

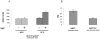


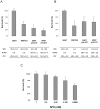
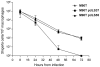
References
-
- Cohen SS. A guide to the polyamines. Oxford Univ. Press, New York, N.Y. USA; 1997.
-
- Seiler N. Functions of polyamine acetylation. Can J Physiol Pharmacol. 1987;65:2024–2035. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

