PLD$ is involved in phagocytosis of microglia: expression and localization changes of PLD4 are correlated with activation state of microglia
- PMID: 22102906
- PMCID: PMC3216956
- DOI: 10.1371/journal.pone.0027544
PLD$ is involved in phagocytosis of microglia: expression and localization changes of PLD4 are correlated with activation state of microglia
Abstract
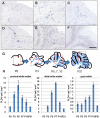
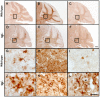
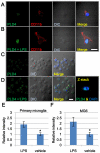
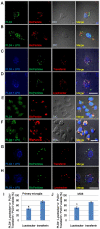
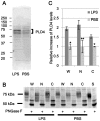
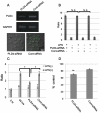
Phospholipase D4 (PLD4) is a recently identified protein that is mainly expressed in the ionized calcium binding adapter molecule 1 (Iba1)-positive microglia in the early postnatal mouse cerebellar white matter. Unlike PLD1 and PLD2, PLD4 exhibits no enzymatic activity for conversion of phosphatidylcholine into choline and phosphatidic acid, and its function is completely unknown. In the present study, we examined the distribution of PLD4 in mouse cerebellar white matter during development and under pathological conditions. Immunohistochemical analysis revealed that PLD4 expression was associated with microglial activation under such two different circumstances. A primary cultured microglia and microglial cell line (MG6) showed that PLD4 was mainly present in the nucleus, except the nucleolus, and expression of PLD4 was upregulated by lipopolysaccharide (LPS) stimulation. In the analysis of phagocytosis of LPS-stimulated microglia, PLD4 was co-localized with phagosomes that contained BioParticles. Inhibition of PLD4 expression using PLD4 specific small interfering RNA (siRNA) in MG6 cells significantly reduced the ratio of phagocytotic cell numbers. These results suggest that the increased PLD4 in the activation process is involved in phagocytosis of activated microglia in the developmental stages and pathological conditions of white matter.
Conflict of interest statement
Figures

References
-
- Pedersen KM, Finsen B, Celis JE, Jensen NA. Expression of a novel murine phospholipase D homolog coincides with late neuronal development in the forebrain. J Biol Chem. 1998;273:31494–31504. - PubMed
-
- Choi SY, Huang P, Jenkins GM, Chan DC, Schiller J, et al. A common lipid links Mfn-mediated mitochondrial fusion and SNARE-regulated exocytosis. Nat Cell Biol. 2006;8:1255–1262. - PubMed
-
- Munck A, Bohm C, Seibel NM, Hashemol HZ, Hampe W. Hu-K4 is a ubiquitously expressed type 2 transmembrane protein associated with the endoplasmic reticulum. FEBS J. 2005;272:1718–1726. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

